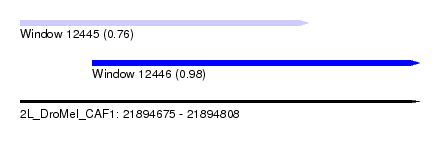
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,894,675 – 21,894,808 |
| Length | 133 |
| Max. P | 0.979890 |

| Location | 21,894,675 – 21,894,771 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.50 |
| Mean single sequence MFE | -34.48 |
| Consensus MFE | -16.10 |
| Energy contribution | -16.93 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.756641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
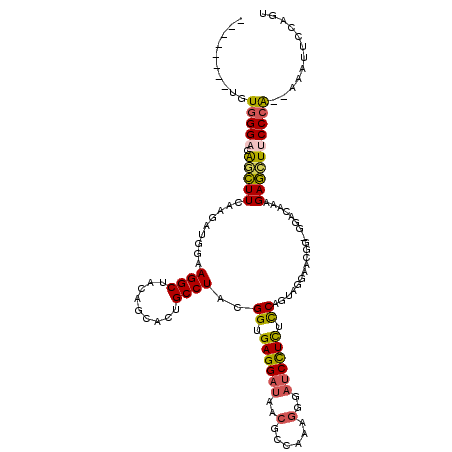
>2L_DroMel_CAF1 21894675 96 + 22407834 UUCU--CUCC-GACGA---GA-AGGCGUGGC-----------------UGUGGGACAGCUUCAAGAUGGAAGGCUACAGCACUGCCUACGGUGAGGAUAACGCCAAAGGUAUCCUCUCCA .(((--(...-...))---))-(((((..((-----------------(((((.....((((......)))).)))))))..)))))..((.(((((((.(......).))))))).)). ( -36.60) >DroSec_CAF1 184510 96 + 1 UUCU--AUCC-AACGA---CA-AGGCGUGGC-----------------UGUGGGACAGCUUCAAGAUUGAAGGCUACAGCACUGCCUACGGUGAGGAUAACGCUAAAGGGAUCCUCUCCA ....--....-.....---..-(((((..((-----------------(((((.....(((((....))))).)))))))..)))))..((.((((((..(......)..)))))).)). ( -34.70) >DroSim_CAF1 128241 96 + 1 UUCU--AUCC-GACGA---CA-AGGCGUGGC-----------------UGUGGGACAGCUUCAAGAUUGAAGGCUACAGCACUGCCUACGGUGAGGAUAACGCUGAAGGGAUCCUCUUCA ...(--((((-.(((.---..-...))).((-----------------((((((.((((((((....)))))((....)).)))))))))))..))))).....((((((...)))))). ( -32.10) >DroEre_CAF1 148487 96 + 1 UUUU--AUCC-GACGA---GA-AAUUCUCUU-----------------UGUGGGAAGGCUUCAAGAUGCAAGGCUACAGCACUGCCUAUGGCGAGGAUAACGCCAAAGGGAUCCUCUCCG ....--.(((-.((((---((-.....)).)-----------------))).)))..((........)).((((.........))))..((.((((((..(......)..)))))).)). ( -28.20) >DroYak_CAF1 142103 96 + 1 UUCU--UUCG-GACGA---GA-AAGACUGGU-----------------UGUGGGACAUCUUCAAGAUGGAAGGCUACAGCACCGCCUAUGGUGAGGAUAACGCCAAAGGGAUCCUCUUCC .(((--(((.-.....---))-))))...((-----------------(((((..((((.....)))).....)))))))((((....))))((((((..(......)..)))))).... ( -29.10) >DroAna_CAF1 128676 117 + 1 GGCAGAACCCAGAGGAUGAAAGUGGCGAGGCUAAAGGUCCCCAUUUCCUUUGGGAGAAGUUCAAAGCCGCAGGCUACAAAACUGCCUACGGAGAGGAUACGGCCCAGAG---CUUUGCCG (((....(((((((((....(((((.(.(((.....)))))))))))))))))).(((((((....(((.((((.........)))).)))...((.......)).)))---))))))). ( -46.20) >consensus UUCU__AUCC_GACGA___GA_AGGCGUGGC_________________UGUGGGACAGCUUCAAGAUGGAAGGCUACAGCACUGCCUACGGUGAGGAUAACGCCAAAGGGAUCCUCUCCA .................................................(((((....((((......))))((....))....)))))((.((((((..(......)..)))))).)). (-16.10 = -16.93 + 0.83)
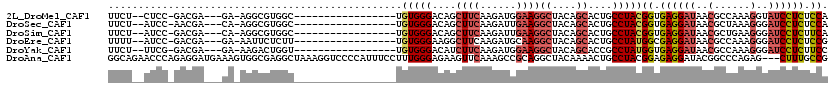
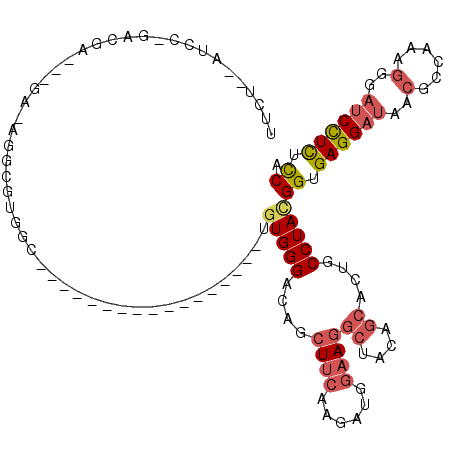
| Location | 21,894,699 – 21,894,808 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.51 |
| Mean single sequence MFE | -36.68 |
| Consensus MFE | -23.66 |
| Energy contribution | -23.55 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21894699 109 + 22407834 --------UGUGGGACAGCUUCAAGAUGGAAGGCUACAGCACUGCCUACGGUGAGGAUAACGCCAAAGGUAUCCUCUCCAAUAGGAACGG-GGAUAAAGAGCUUCCCA--AAAGUCCAGU --------.(((((.(((((((......))))((....)).))))))))((.(((((((.(......).))))))).))....(((...(-(((.........)))).--....)))... ( -37.90) >DroSec_CAF1 184534 109 + 1 --------UGUGGGACAGCUUCAAGAUUGAAGGCUACAGCACUGCCUACGGUGAGGAUAACGCUAAAGGGAUCCUCUCCAGUAGGAACGG-GGACAAAGAGCUUCCCA--AAAUUCCAGU --------.(((((.((((((((....)))))((....)).))))))))((.((((((..(......)..)))))).))....((((..(-(((.........)))).--...))))... ( -38.60) >DroSim_CAF1 128265 109 + 1 --------UGUGGGACAGCUUCAAGAUUGAAGGCUACAGCACUGCCUACGGUGAGGAUAACGCUGAAGGGAUCCUCUUCAGUAGGAACGG-GGACAAAGAGCUUCCCA--AAAUUCCAGU --------.(((((.((((((((....)))))((....)).))))))))((.((((((..(......)..)))))).))....((((..(-(((.........)))).--...))))... ( -35.40) >DroEre_CAF1 148511 109 + 1 --------UGUGGGAAGGCUUCAAGAUGCAAGGCUACAGCACUGCCUAUGGCGAGGAUAACGCCAAAGGGAUCCUCUCCGGAAGGAACGG-GGACGAAGAGCUGCCCA--AAAAUAAAGU --------..((((..(((((.........))))).((((.((.(((.(((((.......))))).)))..((((((((....)))..))-)))...)).))))))))--.......... ( -37.70) >DroYak_CAF1 142127 109 + 1 --------UGUGGGACAUCUUCAAGAUGGAAGGCUACAGCACCGCCUAUGGUGAGGAUAACGCCAAAGGGAUCCUCUUCCGUAGAAAUGG-GCACAAAGAGCUUCCCA--AAAAUAAAGU --------..(((((((((.....))))...((((...((.(((.((((((.((((((..(......)..))))))..))))))...)))-))......)))))))))--.......... ( -36.20) >DroAna_CAF1 128716 114 + 1 CCAUUUCCUUUGGGAGAAGUUCAAAGCCGCAGGCUACAAAACUGCCUACGGAGAGGAUACGGCCCAGAG---CUUUGCCGACAGGACUGAAAGGUGGAGAUUUGCCAGGGAAUUUCC--- ......(((.(.((..((((((....(((.((((.........)))).)))...((.......)).)))---)))..)).).)))(((....)))((((((((......))))))))--- ( -34.30) >consensus ________UGUGGGACAGCUUCAAGAUGGAAGGCUACAGCACUGCCUACGGUGAGGAUAACGCCAAAGGGAUCCUCUCCAGUAGGAACGG_GGACAAAGAGCUUCCCA__AAAUUCCAGU ..........(((((.(((((.........((((.........))))..((.((((((..(......)..)))))).))...................))))))))))............ (-23.66 = -23.55 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:17 2006