
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,861,394 – 21,861,546 |
| Length | 152 |
| Max. P | 0.878729 |

| Location | 21,861,394 – 21,861,507 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.37 |
| Mean single sequence MFE | -29.92 |
| Consensus MFE | -27.24 |
| Energy contribution | -27.35 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21861394 113 + 22407834 AUUGUCACCAUCCACAGUGCUUGCGGCAAAUUAUGGGACAAUAUUUUAGGCUAAUGUUAAUGUUUUCCCAAAAAUGUUCUCAUUGUGCUGAGGAGGGUUCUCCGCUACCCGCU------- ..........(((.(((..(.((.((((.....(((((.((((((...(((....))))))))).)))))....)).)).))..)..))).)))((((........))))...------- ( -28.90) >DroSec_CAF1 149390 113 + 1 AUUGUCACCAUCCACAGUGCUUGCGGCAAAUUAUGGGACAAUAUUUUAGGCUAAUGUUAAUGUUUUCCCAAAAAUGUUCUCAUUGUGCUGAGGAGGGUUCUCCGCUACCCGCU------- ..........(((.(((..(.((.((((.....(((((.((((((...(((....))))))))).)))))....)).)).))..)..))).)))((((........))))...------- ( -28.90) >DroSim_CAF1 95784 120 + 1 AUUGUCACCAUCCACAGUGCUUGCGGCAAAUUAUGGGACAAUAUUUUAGGCUAAUGUUAAUGUUUUCCCAAAAAUGUUCUCAUUGUGCUGAGGAGGGUUCUCCGCUACCCGCUACCCGCU ..........(((.(((..(.((.((((.....(((((.((((((...(((....))))))))).)))))....)).)).))..)..))).)))((((........))))((.....)). ( -29.60) >DroEre_CAF1 111789 106 + 1 AUUGUCACCAUCCACAGUGCUUGCGGCAAAUUAUGGGACAAUAUUUUAGGCUAAUGUUAAUGUUUUCCCAAAAAUGUUCUCAUUGUGCCGAGGAGGGUUCUCCGCU-------------- ((((((.((((......(((.....)))....))))))))))......((((((((..((((((((....))))))))..))))).)))..((((....))))...-------------- ( -30.00) >DroYak_CAF1 103268 113 + 1 AUUGUCACCAUCCGCAGUGCCCGCGGCAAAUUAUGGGACAAUAUUUUAGGCUAAUGUUAAUGUUUUCCCAAAAAUGUUCUCAUUGUGCUGAGGAGGGUUCUCCGUUUCCCUCC------- ((((((.((((((((.......))))......))))))))))......((((((((..((((((((....))))))))..))))).)))..((((((..........))))))------- ( -33.60) >DroAna_CAF1 99098 107 + 1 AUUGUCACCAUCAACAGUGCUCGCGGCAAAUUAUGGGACAAUAUUUUAGGCUAAUGUUAAUGUUUUCCCAAAAAUGUUCUCAUUGUGCCAAGGAGGGACCCACUCUU------------- ((((((.((((......(((.....)))....))))))))))......((((((((..((((((((....))))))))..))))).))).((((((....).)))))------------- ( -28.50) >consensus AUUGUCACCAUCCACAGUGCUUGCGGCAAAUUAUGGGACAAUAUUUUAGGCUAAUGUUAAUGUUUUCCCAAAAAUGUUCUCAUUGUGCUGAGGAGGGUUCUCCGCUACCCGCU_______ ((((((.((((......(((.....)))....))))))))))......((((((((..((((((((....))))))))..))))).)))..((((....))))................. (-27.24 = -27.35 + 0.11)

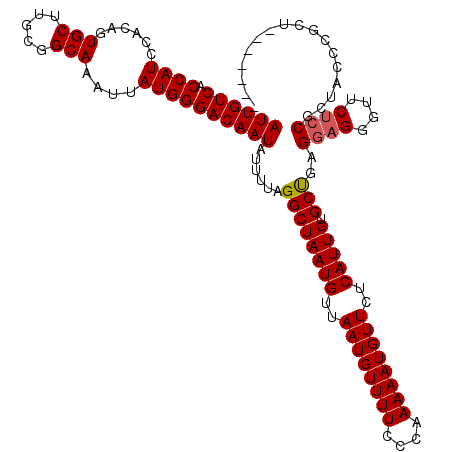

| Location | 21,861,434 – 21,861,546 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 84.91 |
| Mean single sequence MFE | -23.86 |
| Consensus MFE | -15.28 |
| Energy contribution | -15.12 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.596452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21861434 112 + 22407834 AUAUUUUAGGCUAAUGUUAAUGUUUUCCCAAAAAUGUUCUCAUUGUGCUGAGGAGGGUUCUCCGCUACCCGCU-------ACCCGCUACCCGGCACCACCACUUCCCAUCAUCGCACCA .........((.((((..((((((((....))))))))..))))((((((.((.((((........))))((.-------....))..)))))))).................)).... ( -24.80) >DroSec_CAF1 149430 112 + 1 AUAUUUUAGGCUAAUGUUAAUGUUUUCCCAAAAAUGUUCUCAUUGUGCUGAGGAGGGUUCUCCGCUACCCGCU-------ACCCGCUACCCGGCACCACCACUUCACAUCAUCGCACCA .........((.((((..((((((((....))))))))..))))((((((.((.((((........))))((.-------....))..)))))))).................)).... ( -24.80) >DroSim_CAF1 95824 119 + 1 AUAUUUUAGGCUAAUGUUAAUGUUUUCCCAAAAAUGUUCUCAUUGUGCUGAGGAGGGUUCUCCGCUACCCGCUACCCGCUACCCGCUACCCGGCACCACCACUUCACAUCAUCGCACCA .........((.((((..((((((((....))))))))..))))((((((.((.((((........))))((.....)).........)))))))).................)).... ( -24.80) >DroEre_CAF1 111829 97 + 1 AUAUUUUAGGCUAAUGUUAAUGUUUUCCCAAAAAUGUUCUCAUUGUGCCGAGGAGGGUUCUCCGCU--------------ACCCGUCACCC--------CCCUCCCCAUCACCUCACCA ........((((((((..((((((((....))))))))..))))).)))((((.((((....((..--------------...))..))))--------.))))............... ( -23.10) >DroYak_CAF1 103308 104 + 1 AUAUUUUAGGCUAAUGUUAAUGUUUUCCCAAAAAUGUUCUCAUUGUGCUGAGGAGGGUUCUCCGUUUCCCUCC-------ACCCACCACCC--------UUCUCCCCAUCACGGCACCA ........((((((((..((((((((....))))))))..))))).)))(((((((((.....((........-------)).....))))--------)))))............... ( -21.80) >consensus AUAUUUUAGGCUAAUGUUAAUGUUUUCCCAAAAAUGUUCUCAUUGUGCUGAGGAGGGUUCUCCGCUACCCGCU_______ACCCGCUACCCGGCACCACCACUUCCCAUCAUCGCACCA ........((((((((..((((((((....))))))))..))))).)))..((((....))))........................................................ (-15.28 = -15.12 + -0.16)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:43:54 2006