
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,854,521 – 21,854,674 |
| Length | 153 |
| Max. P | 0.998816 |

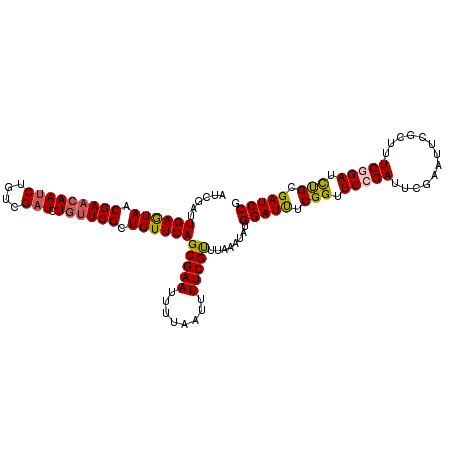
| Location | 21,854,521 – 21,854,634 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 91.24 |
| Mean single sequence MFE | -31.54 |
| Consensus MFE | -28.20 |
| Energy contribution | -28.08 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.24 |
| SVM RNA-class probability | 0.998816 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21854521 113 + 22407834 AUCGAUUGAGUAACGAACAAUCUGUCGAUCUGUUCGCUGUUCAGCGAAUUUUAAUUUUCGUUUAAAUAUGGAUUUCGGUUCCGAUUCGAAUUCUCUUUCGGAUCUGCGAUCCG .(((.(((((((.(((((((((....))).)))))).))))))))))......................(((((.(((.(((((...((....))..))))).))).))))). ( -31.50) >DroSec_CAF1 142316 113 + 1 AUCGAUUGAGUAACGAACAAUCUGUCGAUCUGCUCGCUGUUCAGCGAAUUUUAAUUUUCGCUUAAAUAUGGAUCUCGGUUCAGAUUCGAAUUCGCUUUCGGAUCUGCGAUCCG ...((.((((((.(((........)))...))))))....))((((((........)))))).......(((((......((((((((((......)))))))))).))))). ( -30.60) >DroSim_CAF1 88615 113 + 1 AUCGAUUGAGUAACGAACAAUCUGUCGAUCUGUUCGCUGUUCAGCGAAUUUUAAUUUUCGCUUAAAUAUGGAUCUCGGUUCCGAUUCGAAUUCGCUUUCGGAUCUGCGAUCCG ......((((((.(((((((((....))).)))))).))))))(((((........)))))........(((((.(((.(((((..((....))...))))).))).))))). ( -37.10) >DroEre_CAF1 104400 113 + 1 AUCGAUUGAGUAACGAACAAUCUGUCGCUCUGUUCGCUGUUCAGCGAAUUUUAAUUUUCGUCCAAAUAUGGAUUUCGGUUCCGAUUUGGAUUCGCUUUCGGAUUCGCGAUCCG ...(((((((((.((((((...........)))))).))))).(((((((..((.....((((((((.((((.......)))))))))))).....))..))))))))))).. ( -31.30) >DroYak_CAF1 94269 109 + 1 AUCGAUUGAAUAACGAAAAAUCUGUCGAUCUGUUCGCUGUUCAGCGAAUUU----UUUCGUUUAAAUAUGGAUUUCGGUUCCGAUUUAGACUCACUUUCAGAUUCGCGAUCCG ...(((((.....(((...(((((..((...(((((((....)))))))..----..))((((((((.((((.......)))))))))))).......))))))))))))).. ( -27.20) >consensus AUCGAUUGAGUAACGAACAAUCUGUCGAUCUGUUCGCUGUUCAGCGAAUUUUAAUUUUCGUUUAAAUAUGGAUUUCGGUUCCGAUUCGAAUUCGCUUUCGGAUCUGCGAUCCG ......((((((.(((((((((....))).)))))).))))))(((((........)))))........(((((.(((.(((((.............))))).))).))))). (-28.20 = -28.08 + -0.12)

| Location | 21,854,521 – 21,854,634 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 91.24 |
| Mean single sequence MFE | -30.70 |
| Consensus MFE | -22.32 |
| Energy contribution | -22.52 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.917974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21854521 113 - 22407834 CGGAUCGCAGAUCCGAAAGAGAAUUCGAAUCGGAACCGAAAUCCAUAUUUAAACGAAAAUUAAAAUUCGCUGAACAGCGAACAGAUCGACAGAUUGUUCGUUACUCAAUCGAU ((((((...)))))).........((((.(((....)))............((((((........((((((....))))))..((((....)))).))))))......)))). ( -27.40) >DroSec_CAF1 142316 113 - 1 CGGAUCGCAGAUCCGAAAGCGAAUUCGAAUCUGAACCGAGAUCCAUAUUUAAGCGAAAAUUAAAAUUCGCUGAACAGCGAGCAGAUCGACAGAUUGUUCGUUACUCAAUCGAU .(((((.(((((.((((......)))).)))))......))))).......((((((........))))))((..(((((((((.((....)))))))))))..))....... ( -34.10) >DroSim_CAF1 88615 113 - 1 CGGAUCGCAGAUCCGAAAGCGAAUUCGAAUCGGAACCGAGAUCCAUAUUUAAGCGAAAAUUAAAAUUCGCUGAACAGCGAACAGAUCGACAGAUUGUUCGUUACUCAAUCGAU .(((((.(.(.(((((...((....))..))))).).).))))).......((((((........))))))((..(((((((((.((....)))))))))))..))....... ( -35.50) >DroEre_CAF1 104400 113 - 1 CGGAUCGCGAAUCCGAAAGCGAAUCCAAAUCGGAACCGAAAUCCAUAUUUGGACGAAAAUUAAAAUUCGCUGAACAGCGAACAGAGCGACAGAUUGUUCGUUACUCAAUCGAU .(((((((..........))).))))...(((....)))..((((....))))(((.........((((((....))))))..(((..((.((....))))..)))..))).. ( -30.70) >DroYak_CAF1 94269 109 - 1 CGGAUCGCGAAUCUGAAAGUGAGUCUAAAUCGGAACCGAAAUCCAUAUUUAAACGAAA----AAAUUCGCUGAACAGCGAACAGAUCGACAGAUUUUUCGUUAUUCAAUCGAU .((((..((..(((((.((.....))...)))))..))..)))).......(((((((----((.((((((....))))))....((....)))))))))))........... ( -25.80) >consensus CGGAUCGCAGAUCCGAAAGCGAAUUCGAAUCGGAACCGAAAUCCAUAUUUAAACGAAAAUUAAAAUUCGCUGAACAGCGAACAGAUCGACAGAUUGUUCGUUACUCAAUCGAU .((((..(.(.(((((...((....))..))))).).)..)))).......((((((........((((((....))))))..((((....)))).))))))........... (-22.32 = -22.52 + 0.20)


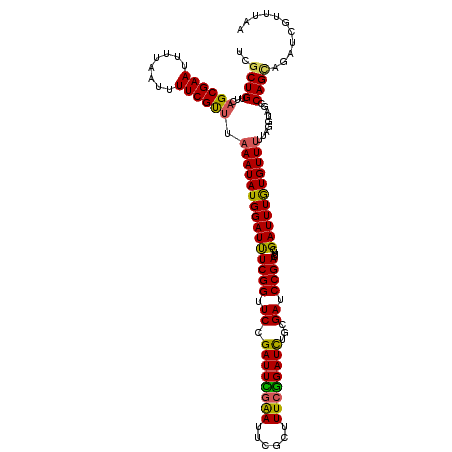
| Location | 21,854,554 – 21,854,674 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.00 |
| Mean single sequence MFE | -31.70 |
| Consensus MFE | -27.32 |
| Energy contribution | -26.48 |
| Covariance contribution | -0.84 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21854554 120 + 22407834 UCGCUGUUCAGCGAAUUUUAAUUUUCGUUUAAAUAUGGAUUUCGGUUCCGAUUCGAAUUCUCUUUCGGAUCUGCGAUCCGACUUCGAUUUGUGUUUUUAGGUAGCCAGCAGAUCGUUUAA (((((....)))))..............((((((...((..((((.(((((((((((......)))))))).).)).))))..))((((((((((.......)))..))))))))))))) ( -31.90) >DroSec_CAF1 142349 120 + 1 UCGCUGUUCAGCGAAUUUUAAUUUUCGCUUAAAUAUGGAUCUCGGUUCAGAUUCGAAUUCGCUUUCGGAUCUGCGAUCCGACUUCGAUUUGUGUUUUUAGGUAGCCAGCAGAUCGUUUAA ..((((...((((((........)))))).(((((..((((((((..((((((((((......))))))))))....))))....))))..))))).........))))........... ( -37.40) >DroSim_CAF1 88648 120 + 1 UCGCUGUUCAGCGAAUUUUAAUUUUCGCUUAAAUAUGGAUCUCGGUUCCGAUUCGAAUUCGCUUUCGGAUCUGCGAUCCGACUUCGAUUUGUGUUUUUAGGUAGCCAGCAGAUCGUUUAA .........((((((........))))))......((((((.(((.(((((..((....))...))))).))).))))))....(((((((((((.......)))..))))))))..... ( -35.20) >DroEre_CAF1 104433 120 + 1 UCGCUGUUCAGCGAAUUUUAAUUUUCGUCCAAAUAUGGAUUUCGGUUCCGAUUUGGAUUCGCUUUCGGAUUCGCGAUCCGACUUCGAUUUGUGUUUUUAGGUAGUCAGUAGAUCGUUUAA (((((....)))))............((((((((.((((.......))))))))))))(((...(((((((...)))))))...))).........(((((..(((....)))..))))) ( -29.90) >DroYak_CAF1 94302 115 + 1 UCGCUGUUCAGCGAAUUU----UUUCGUUUAAAUAUGGAUUUCGGUUCCGAUUUAGACUCACUUUCAGAUUCGCGAUCCGAAUUCGAUUUAUGUU-UUAGGUAGUCAGAAGAUCGUUUAA (((((....)))))....----........(((((((((((((((.(((((.((.((.......)).)).))).)).))))....))))))))))-)((((..(((....)))..)))). ( -24.10) >consensus UCGCUGUUCAGCGAAUUUUAAUUUUCGUUUAAAUAUGGAUUUCGGUUCCGAUUCGAAUUCGCUUUCGGAUCUGCGAUCCGACUUCGAUUUGUGUUUUUAGGUAGCCAGCAGAUCGUUUAA ..((((...((((((........)))))).(((((((((((((((.((.((((((((......))))))))...)).))))....))))))))))).........))))........... (-27.32 = -26.48 + -0.84)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:43:47 2006