
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,849,288 – 21,849,475 |
| Length | 187 |
| Max. P | 0.915422 |

| Location | 21,849,288 – 21,849,384 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 81.60 |
| Mean single sequence MFE | -22.72 |
| Consensus MFE | -16.20 |
| Energy contribution | -17.82 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915422 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
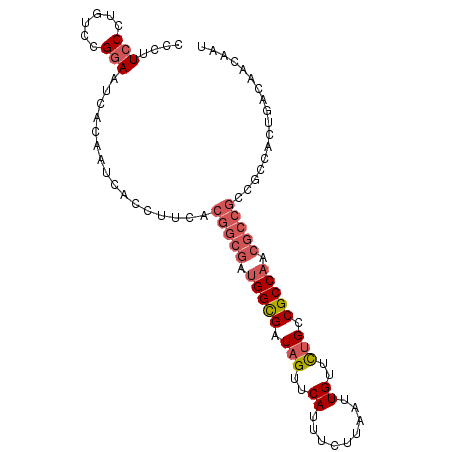
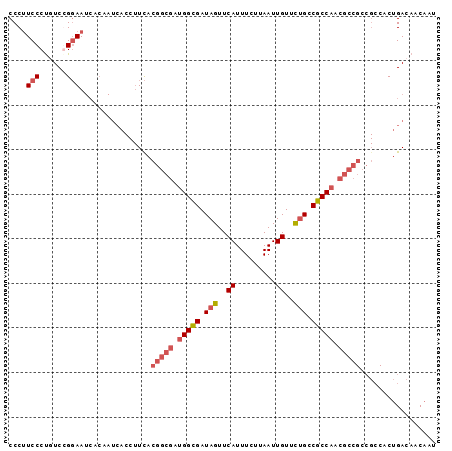
>2L_DroMel_CAF1 21849288 96 - 22407834 CCCUUCCCCUUCGGCAAUCACGAUCACCUUUACGGCGAUGGCGAUAGUUCAUUUCUUAAUUGUUCUGCCGCCAACGCCGCCGCCACUGACAACAAU ............(((......((......)).(((((.(((((.(((..((.........))..))).))))).)))))..)))............ ( -25.40) >DroSec_CAF1 137149 96 - 1 CCCUUCCCUGUCCGGAAUCACAAUCACCUUCACGGCGAUGGCGAUAGUUCAUUUCUUAAUUGUUCUGCCGCCAACGCCGCCGCCACUGACAACAAU ........((((.((((...........))).(((((.(((((.(((..((.........))..))).))))).)))))......).))))..... ( -24.40) >DroSim_CAF1 83590 96 - 1 CCCUUCCCUGUCCGGAAUCACAAUCACCUUCACGGCGAUGGCGAUAGUUCAUUUCUUAAUUGUUCUGCCGCCAACGCCGCCGCCACUGACAACAAU ........((((.((((...........))).(((((.(((((.(((..((.........))..))).))))).)))))......).))))..... ( -24.40) >DroYak_CAF1 88158 72 - 1 CCCAUCCCUUUCGGGAA------------UCACGGCGAUGGUGAUAGUUCAUUUCUUAAUUGUUUGGCCGCC------------ACGGACAACAAU .((.((((....)))).------------....((((.....(((((((........)))))))....))))------------..))........ ( -16.70) >consensus CCCUUCCCUGUCCGGAAUCACAAUCACCUUCACGGCGAUGGCGAUAGUUCAUUUCUUAAUUGUUCUGCCGCCAACGCCGCCGCCACUGACAACAAU ....(((......)))................(((((.(((((.(((..((.........))..))).))))).)))))................. (-16.20 = -17.82 + 1.62)

| Location | 21,849,384 – 21,849,475 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 90.62 |
| Mean single sequence MFE | -32.18 |
| Consensus MFE | -27.12 |
| Energy contribution | -27.32 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
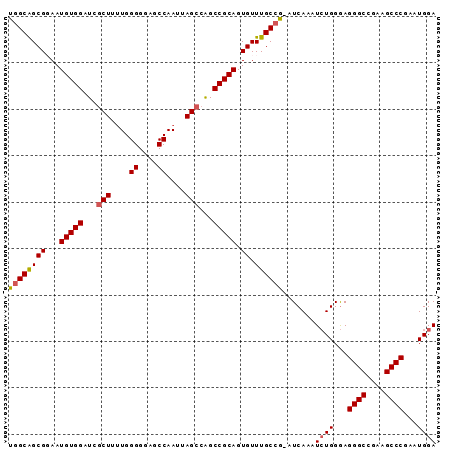
>2L_DroMel_CAF1 21849384 91 + 22407834 UGGCAGCGGAAUGUGGAUCUCUUGUGGGGGAGCCAAUUAGCCAGCCGCAGUGUUUGCCG-AUCAAAUCUGGGAGGGCCGAAGCCCGAAUGGA .(((..((.....(((.(((((....))))).)))....(((..((.(((.(((((...-..))))))))))..)))))..)))........ ( -27.70) >DroSec_CAF1 137245 92 + 1 UGGCAGCGCAAUGUGGAUCGCUAUUGGGGGAGCCAAUUAGCCAGCCGCAGUGUUUGCCAAAUCAAAUCUGGGAGGGCCGUUGCCCAAAUGGA ((((((.(((.(((((...((((...((....))...))))...))))).))))))))).......((((...((((....))))...)))) ( -33.90) >DroSim_CAF1 83686 92 + 1 UGGCAGCGCAAUGUGGAUCGCUAUUGGGGGAGCCAAUUAGCCAGCCGCAGUGUUUGCCAAAUCAAAUCUGGGAGGGCCGUUGCCCGAAUGGA ((((((.(((.(((((...((((...((....))...))))...))))).))))))))).......((((...((((....))))...)))) ( -35.00) >DroEre_CAF1 99065 91 + 1 UGGCGGCGGAAUGUGGAUCGCUUUUGGGGAAGCCAAUUAGCCAGCCGCAGUGUUUGCCG-AUCAAAUCUGGGAGGGCUGAAGCCCAAAUGGA ((((((((((.(((((...(((.((((.....))))..)))...)))))...)))))))-.)))..((((...((((....))))...)))) ( -34.80) >DroYak_CAF1 88230 91 + 1 UUGCAGCGGAAUGUGGAUCGCUUUUGGGGAAACCAAUUAGCCGGCCGCAGUGUUUGCCG-AUCAAAUAUGGAAGGGCUGAAGCCCGAAUGGA .....(((((.(((((...(((....((....))....)))...)))))...)))))..-.............((((....))))....... ( -29.50) >consensus UGGCAGCGGAAUGUGGAUCGCUUUUGGGGGAGCCAAUUAGCCAGCCGCAGUGUUUGCCG_AUCAAAUCUGGGAGGGCCGAAGCCCGAAUGGA ((((((((...(((((...(((....((....))....)))...))))).)).)))))).......((((...((((....))))...)))) (-27.12 = -27.32 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:43:42 2006