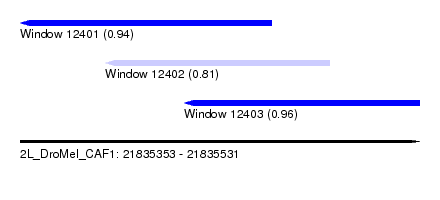
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,835,353 – 21,835,531 |
| Length | 178 |
| Max. P | 0.956760 |

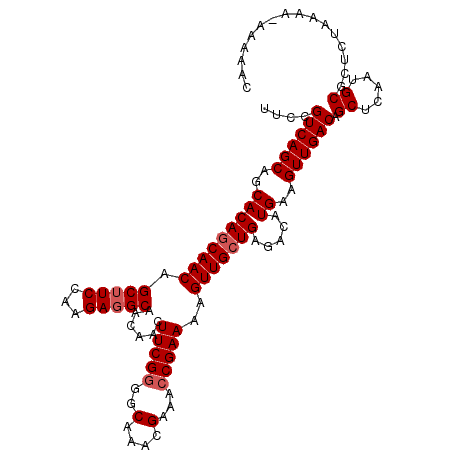
| Location | 21,835,353 – 21,835,465 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 96.41 |
| Mean single sequence MFE | -29.42 |
| Consensus MFE | -26.85 |
| Energy contribution | -27.60 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.937915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21835353 112 - 22407834 UUCCGUCAGCAGCACAACAACAGCUUCCAAGAGGCAACAACUUCGGGGCAAACGAAACCGAAAAGUUGCUGAGACAUGAAGUUGACAGCUCAAUGCGCUCGAAAAAAAAAAC .....(((((..........(((((((.....((((((...(((((...........)))))..)))))).......)))))))...((.....))))).)).......... ( -27.20) >DroSec_CAF1 122622 111 - 1 UUCCGUCAGCAGCACAGCAACAGCUUCCAAGAGGCAACAACUUCGGGGCAAACGAAACCGAAAAGUUGCUGAGACGUGAAGUUGACAGCUCAAUGCGCUCUAAAA-AAAAAC ....((((((..(((((((((.(((((...)))))......(((((...........)))))..)))))).....)))..)))))).((.....)).........-...... ( -30.80) >DroSim_CAF1 69895 111 - 1 UUCCGUCAGCAGCACAGCAACAGCUUCCAAGAGGCAACAACUUCGGGGCAAACGAAACCGAAAAGUUGCUGAGACGUGAAGUUGACAGCUCAAUGCGCUCUAAAA-AAAAAC ....((((((..(((((((((.(((((...)))))......(((((...........)))))..)))))).....)))..)))))).((.....)).........-...... ( -30.80) >DroEre_CAF1 83899 110 - 1 UUCCGUCAGCAGCACAGCAACAGCGUCCAAGAGGCAACAACUUCGGGGCAAACGAAAACGAAAAGUUGCUGAGACAUGAAGUUGACAGCUCAAUGCGCUCCAAAA--AAAAC ....((((((..(((((((((...((((..(((.......)))..))))...((....))....))))))).....))..)))))).((.....)).........--..... ( -28.90) >consensus UUCCGUCAGCAGCACAGCAACAGCUUCCAAGAGGCAACAACUUCGGGGCAAACGAAACCGAAAAGUUGCUGAGACAUGAAGUUGACAGCUCAAUGCGCUCUAAAA_AAAAAC ....((((((..(((((((((.(((((...)))))......(((((..(....)...)))))..))))))).....))..)))))).((.....))................ (-26.85 = -27.60 + 0.75)


| Location | 21,835,391 – 21,835,491 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 97.20 |
| Mean single sequence MFE | -30.60 |
| Consensus MFE | -26.44 |
| Energy contribution | -26.52 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814098 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21835391 100 - 22407834 CCUGAUUGGUCUUUGCUCUGGCCAGAUUCCGUCAGCAGCACAACAACAGCUUCCAAGAGGCAACAACUUCGGGGCAAACGAAACCGAAAAGUUGCUGAGA .....((((((........))))))......((((((((.........(((((...)))))......(((((...........)))))..)))))))).. ( -29.90) >DroSec_CAF1 122659 100 - 1 CCUGAUUGGUCUUUGCUCUGGCCAGAUUCCGUCAGCAGCACAGCAACAGCUUCCAAGAGGCAACAACUUCGGGGCAAACGAAACCGAAAAGUUGCUGAGA ((.....))....(((((((((........))))).))))(((((((.(((((...)))))......(((((...........)))))..)))))))... ( -31.80) >DroSim_CAF1 69932 100 - 1 CCUGAUUGGUCUUUGCUCUGGCCAGAUUCCGUCAGCAGCACAGCAACAGCUUCCAAGAGGCAACAACUUCGGGGCAAACGAAACCGAAAAGUUGCUGAGA ((.....))....(((((((((........))))).))))(((((((.(((((...)))))......(((((...........)))))..)))))))... ( -31.80) >DroEre_CAF1 83935 100 - 1 CCUGAUUGGUCUUUGCUCUGGCCAGAUUCCGUCAGCAGCACAGCAACAGCGUCCAAGAGGCAACAACUUCGGGGCAAACGAAAACGAAAAGUUGCUGAGA ((.....))....(((((((((........))))).))))(((((((...((((..(((.......)))..))))...((....))....)))))))... ( -30.00) >DroYak_CAF1 74832 100 - 1 CCUGAUUGGUCUUUGCUUUGGCCAGAUUCCGUCAGCAGCACAGCAACAGCGCCCAAGAGGCAACAACAUCGGGGCAAACGAAAACGAAAAGUUGCUGAGA .....((((((........))))))......((((((((...((....))((((....(....).......))))...............)))))))).. ( -29.50) >consensus CCUGAUUGGUCUUUGCUCUGGCCAGAUUCCGUCAGCAGCACAGCAACAGCUUCCAAGAGGCAACAACUUCGGGGCAAACGAAACCGAAAAGUUGCUGAGA ((.....))....(((((((((........))))).))))(((((((.((.((((((.(....)..))).)))))...((....))....)))))))... (-26.44 = -26.52 + 0.08)

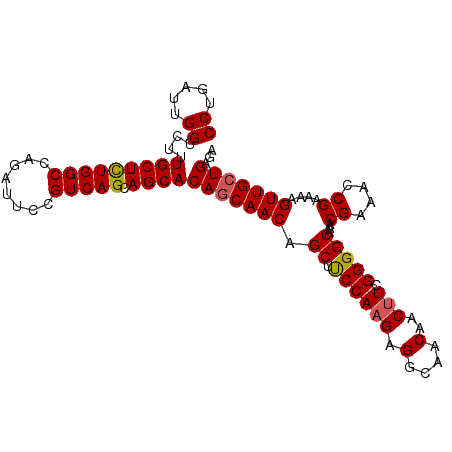
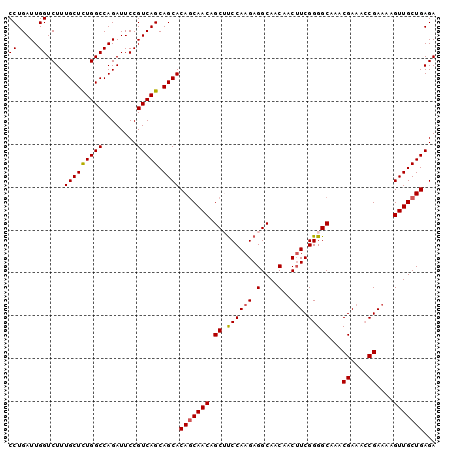
| Location | 21,835,426 – 21,835,531 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 97.52 |
| Mean single sequence MFE | -34.32 |
| Consensus MFE | -32.28 |
| Energy contribution | -32.92 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21835426 105 - 22407834 CAAGUGGAAAUCCUUUUGGCCGAAAGGAGGCAGCAACCGACCUGAUUGGUCUUUGCUCUGGCCAGAUUCCGUCAGCAGCACAACAACAGCUUCCAAGAGGCAACA ((((.((....)).))))(((....((((((.......((((.....))))..(((((((((........))))).))))........))))))....))).... ( -35.60) >DroSec_CAF1 122694 105 - 1 CAAGUGGAAAUCCUUUUGGCCGAAAGGAGGCAGCAACCGACCUGAUUGGUCUUUGCUCUGGCCAGAUUCCGUCAGCAGCACAGCAACAGCUUCCAAGAGGCAACA ((((.((....)).))))(((....((((((.((....((((.....))))..(((((((((........))))).))))..))....))))))....))).... ( -35.70) >DroSim_CAF1 69967 105 - 1 CAAGUGGAAAUCCUUUUGGCCGAAAGGAGGCAGCAACCGACCUGAUUGGUCUUUGCUCUGGCCAGAUUCCGUCAGCAGCACAGCAACAGCUUCCAAGAGGCAACA ((((.((....)).))))(((....((((((.((....((((.....))))..(((((((((........))))).))))..))....))))))....))).... ( -35.70) >DroEre_CAF1 83970 105 - 1 CAAGUGGAAAUCCUUUUGGCCGAAAGGAGGCAGCAACCGACCUGAUUGGUCUUUGCUCUGGCCAGAUUCCGUCAGCAGCACAGCAACAGCGUCCAAGAGGCAACA ((((.((....)).))))(((....(((.((.((....((((.....))))..(((((((((........))))).))))..))....)).)))....))).... ( -34.00) >DroYak_CAF1 74867 105 - 1 CAAGUGGAAAUCCUUUAGGCCGGAAGGAGGCAGCAACCGACCUGAUUGGUCUUUGCUUUGGCCAGAUUCCGUCAGCAGCACAGCAACAGCGCCCAAGAGGCAACA ...........(((((.((((((((...((((((((..((((.....)))).)))))...)))...)))))...((......))......)))..)))))..... ( -30.60) >consensus CAAGUGGAAAUCCUUUUGGCCGAAAGGAGGCAGCAACCGACCUGAUUGGUCUUUGCUCUGGCCAGAUUCCGUCAGCAGCACAGCAACAGCUUCCAAGAGGCAACA ((((.((....)).))))(((....((((((.......((((.....))))..(((((((((........))))).))))........))))))....))).... (-32.28 = -32.92 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:43:36 2006