
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,829,844 – 21,829,956 |
| Length | 112 |
| Max. P | 0.882083 |

| Location | 21,829,844 – 21,829,956 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 82.80 |
| Mean single sequence MFE | -50.33 |
| Consensus MFE | -42.70 |
| Energy contribution | -42.27 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.700239 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
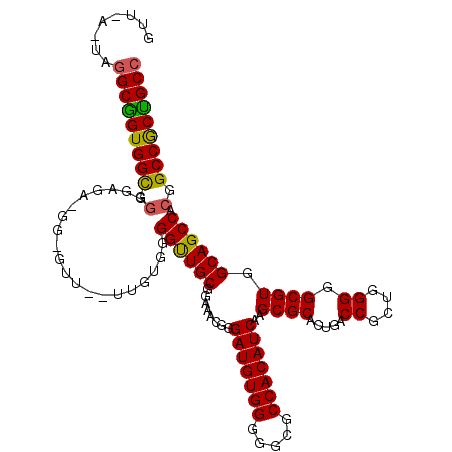
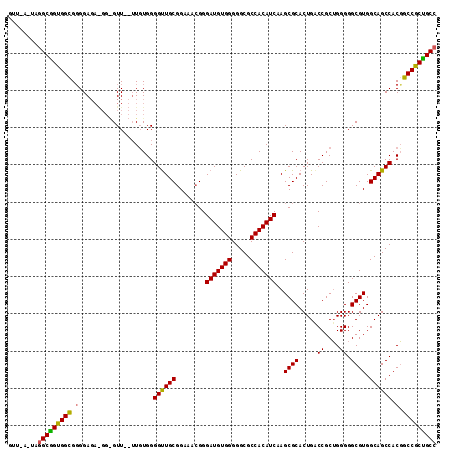
>2L_DroMel_CAF1 21829844 112 + 22407834 GUU---UGAGCGGUGGUGGGGAGAGGGAGUUAUAUGUGGGGUUGCGGAAACGGGAUGUGGGGGCGCCACAUCAAGCGCACUGGCCGCUGGGGGCGUGGCAGCCACGGCCGCCGCC ...---...((((((((.(((..............(((.(.((.((....)).(((((((.....))))))))).).)))..(((((.......)))))..)).).)))))))). ( -47.50) >DroEre_CAF1 78179 113 + 1 GUUCACUUGGCGGUGGCGGGGAGAAGGGGUU--UUGUGGGGCUGCGGAAAAGGGAUGUGGGGGCGCCACAUCAAGCGCACUGACCGCAGGGGGCGUGGCAGCCACUGCCACUGCC ........(((((((((((.((((.....))--))....((((((........(((((((.....)))))))..((((.(((....)))...)))).)))))).))))))))))) ( -52.40) >DroYak_CAF1 67811 107 + 1 UGUGAAUAGGCAGUGGCGUGGA------GUU--UUCUGGGGUUGCGGAAACGGGAUGUGGGGGCGCCACAUCAAGCGCACUGACCGCUGGGGGCGUGGCAGCCACGGCCGCUGCC ........(((((((((((((.------(((--..(..(.(((.((....)).))).))..)))(((((..(.((((.......))))...)..)))))..)))).))))))))) ( -51.10) >consensus GUU_A_UAGGCGGUGGCGGGGAGA_GG_GUU__UUGUGGGGUUGCGGAAACGGGAUGUGGGGGCGCCACAUCAAGCGCACUGACCGCUGGGGGCGUGGCAGCCACGGCCGCUGCC ........(((((((((.(....................((((((........(((((((.....)))))))..((((.....((....)).)))).)))))).).))))))))) (-42.70 = -42.27 + -0.44)

| Location | 21,829,844 – 21,829,956 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 82.80 |
| Mean single sequence MFE | -37.33 |
| Consensus MFE | -33.43 |
| Energy contribution | -33.43 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
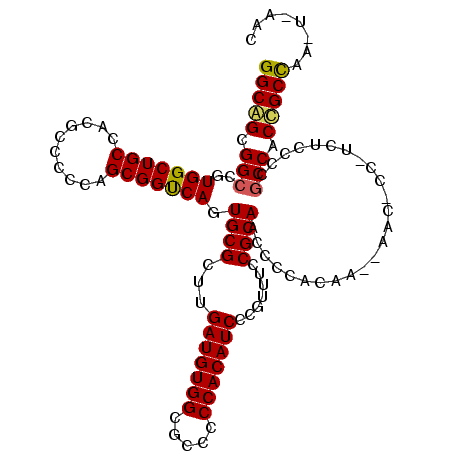
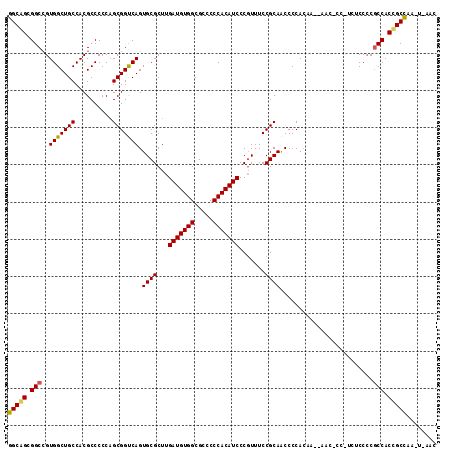
>2L_DroMel_CAF1 21829844 112 - 22407834 GGCGGCGGCCGUGGCUGCCACGCCCCCAGCGGCCAGUGCGCUUGAUGUGGCGCCCCCACAUCCCGUUUCCGCAACCCCACAUAUAACUCCCUCUCCCCACCACCGCUCA---AAC (((((.(((((((((......)))....))))))..((((...(((((((.....))))))).......)))).............................)))))..---... ( -34.80) >DroEre_CAF1 78179 113 - 1 GGCAGUGGCAGUGGCUGCCACGCCCCCUGCGGUCAGUGCGCUUGAUGUGGCGCCCCCACAUCCCUUUUCCGCAGCCCCACAA--AACCCCUUCUCCCCGCCACCGCCAAGUGAAC (((.(((((.((((...)))).....((((((..((....)).(((((((.....)))))))......))))))........--..............))))).)))........ ( -38.00) >DroYak_CAF1 67811 107 - 1 GGCAGCGGCCGUGGCUGCCACGCCCCCAGCGGUCAGUGCGCUUGAUGUGGCGCCCCCACAUCCCGUUUCCGCAACCCCAGAA--AAC------UCCACGCCACUGCCUAUUCACA (((((.((((((((...)))))......((((.....(((...(((((((.....))))))).)))..))))..........--...------.....))).)))))........ ( -39.20) >consensus GGCAGCGGCCGUGGCUGCCACGCCCCCAGCGGUCAGUGCGCUUGAUGUGGCGCCCCCACAUCCCGUUUCCGCAACCCCACAA__AAC_CC_UCUCCCCGCCACCGCCAA_U_AAC (((((.(((..(((((((..........))))))).((((...(((((((.....))))))).......)))).........................))).)))))........ (-33.43 = -33.43 + 0.01)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:43:29 2006