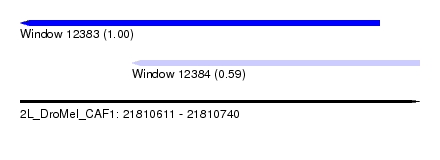
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,810,611 – 21,810,740 |
| Length | 129 |
| Max. P | 0.996705 |

| Location | 21,810,611 – 21,810,727 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.09 |
| Mean single sequence MFE | -30.64 |
| Consensus MFE | -28.12 |
| Energy contribution | -27.92 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.996705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
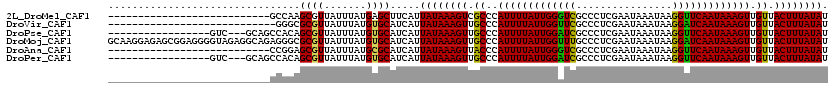
>2L_DroMel_CAF1 21810611 116 - 22407834 UAUGAGCUUCAUUAUAAAGUCGCCCAUUUUAUUGGGUCGCCCUCGAAUAAAUAAGGUUCAAUAAAGUUGUUACUUUAUAUUGACACUCGCAGGCGGCAAUCGCUCGUCACAUUCUG---- ...(((..(((.((((((((.((..(((((((((..((................))..))))))))).)).)))))))).)))..)))...((((((....)).))))........---- ( -31.59) >DroVir_CAF1 66146 117 - 1 UAUGUGCAUCAUUAUAAAGUUGCCCAUUUUAUUGGUUCGCCCUCGAAUAAAUAAGGAUCAAUAAAGUUGUUACUUUAUAUUGACACUUGCAGGCGGCAAUCGCUCGUCACUUUCGGU--- ....(((((((.((((((((.((..(((((((((((((................))))))))))))).)).)))))))).)))....))))((((((....)).)))).........--- ( -31.59) >DroPse_CAF1 72657 120 - 1 UAUGUGCAUCAUUAUAAAGUUGCCCAUUUUAUUGGAUCGCCCUCGAAUAAAUAAGGUUCAAUAAAGUUGUUACUUUAUAUUGACACUUGCAGGCGGCAAUCGCUCGUCACUUUCGGUUGU ....(((((((.((((((((.((..(((((((((((((................))))))))))))).)).)))))))).)))....))))((((((....)).))))............ ( -31.59) >DroYak_CAF1 47022 116 - 1 UAUGAGAAUCAUUAUAAAGUCGCCCAUUUUAUUGGGUCGCCCUCGAAUAAAUAAGGUUCAAUAAAGUUGUUACUUUAUAUUGACACGCGCAGGCGGCAAUCGCUCGUCACAUUCGG---- .....((((((.((((((((.((..(((((((((..((................))..))))))))).)).)))))))).)))........((((((....)).))))...)))..---- ( -28.29) >DroMoj_CAF1 69915 116 - 1 UAUGUGCAUCAUUAUAAAGUUGCCCAUUUUAUUGGUUUGCCCUCGAAUAAAUAAGGAUCAAUAAAGUUGUUACUUUAUAUUGACACUUGCAGGCGGCAAUCGCUCGUCACUCUCA-U--- ....(((((((.((((((((.((..(((((((((((((................))))))))))))).)).)))))))).)))....))))((((((....)).)))).......-.--- ( -29.19) >DroPer_CAF1 74606 120 - 1 UAUGUGCAUCAUUAUAAAGUUGCCCAUUUUAUUGGAUCGCCCUCGAAUAAAUAAGGUUCAAUAAAGUUGUUACUUUAUAUUGACACUUGCAGGCGGCAAUCGCUCGUCACUUUCGGUUGU ....(((((((.((((((((.((..(((((((((((((................))))))))))))).)).)))))))).)))....))))((((((....)).))))............ ( -31.59) >consensus UAUGUGCAUCAUUAUAAAGUUGCCCAUUUUAUUGGAUCGCCCUCGAAUAAAUAAGGUUCAAUAAAGUUGUUACUUUAUAUUGACACUUGCAGGCGGCAAUCGCUCGUCACUUUCGGU___ .....((((((.((((((((.((..(((((((((((((................))))))))))))).)).)))))))).)))....))).((((((....)).))))............ (-28.12 = -27.92 + -0.19)


| Location | 21,810,647 – 21,810,740 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.28 |
| Mean single sequence MFE | -22.40 |
| Consensus MFE | -18.17 |
| Energy contribution | -17.39 |
| Covariance contribution | -0.78 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21810647 93 - 22407834 ---------------------------GCCAAGCGUUAUUUAUGAGCUUCAUUAUAAAGUCGCCCAUUUUAUUGGGUCGCCCUCGAAUAAAUAAGGUUCAAUAAAGUUGUUACUUUAUAU ---------------------------....(((.(((((((((((............((.(((((......))))).)).)))..)))))))).)))..(((((((....))))))).. ( -18.82) >DroVir_CAF1 66183 92 - 1 ----------------------------GGGCGCGUUAUUUAUGUGCAUCAUUAUAAAGUUGCCCAUUUUAUUGGUUCGCCCUCGAAUAAAUAAGGAUCAAUAAAGUUGUUACUUUAUAU ----------------------------..((((((.....)))))).....((((((((.((..(((((((((((((................))))))))))))).)).)))))))). ( -24.99) >DroPse_CAF1 72697 100 - 1 -----------------GUC---GCAGCCACAGCGUUAUUUAUGUGCAUCAUUAUAAAGUUGCCCAUUUUAUUGGAUCGCCCUCGAAUAAAUAAGGUUCAAUAAAGUUGUUACUUUAUAU -----------------...---...((.(((..........))))).....((((((((.((..(((((((((((((................))))))))))))).)).)))))))). ( -20.49) >DroMoj_CAF1 69951 120 - 1 GCAAGGAGAGCGGAGGGGUAGAGGCAGAGGGCGCGUUAUUUAUGUGCAUCAUUAUAAAGUUGCCCAUUUUAUUGGUUUGCCCUCGAAUAAAUAAGGAUCAAUAAAGUUGUUACUUUAUAU .....(((.(((((..(((((((((((...((((((.....))))))............)))))...))))))..))))).)))................(((((((....))))))).. ( -26.96) >DroAna_CAF1 50884 93 - 1 ---------------------------CCGGAGCGUUAUUUAUGCGCAUCAUUAUAAAGUUACCCAUUUUAUUGGGUCGCCCUCGAAUAAAUAAGGUUCAAUAAAGUUGUUACUUUAUAU ---------------------------...((((.((((((((.((............((.(((((......))))).))...)).)))))))).)))).(((((((....))))))).. ( -22.66) >DroPer_CAF1 74646 100 - 1 -----------------GUC---GCAGCCACAGCGUUAUUUAUGUGCAUCAUUAUAAAGUUGCCCAUUUUAUUGGAUCGCCCUCGAAUAAAUAAGGUUCAAUAAAGUUGUUACUUUAUAU -----------------...---...((.(((..........))))).....((((((((.((..(((((((((((((................))))))))))))).)).)))))))). ( -20.49) >consensus ___________________________CCGGAGCGUUAUUUAUGUGCAUCAUUAUAAAGUUGCCCAUUUUAUUGGAUCGCCCUCGAAUAAAUAAGGUUCAAUAAAGUUGUUACUUUAUAU ................................((((.......)))).....((((((((.((..(((((((((((((................))))))))))))).)).)))))))). (-18.17 = -17.39 + -0.78)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:43:19 2006