| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,196,967 – 2,197,059 |
| Length | 92 |
| Max. P | 0.591624 |

| Location | 2,196,967 – 2,197,059 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 74.62 |
| Mean single sequence MFE | -20.25 |
| Consensus MFE | -14.14 |
| Energy contribution | -14.00 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.11 |
| Mean z-score | -0.94 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.591624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
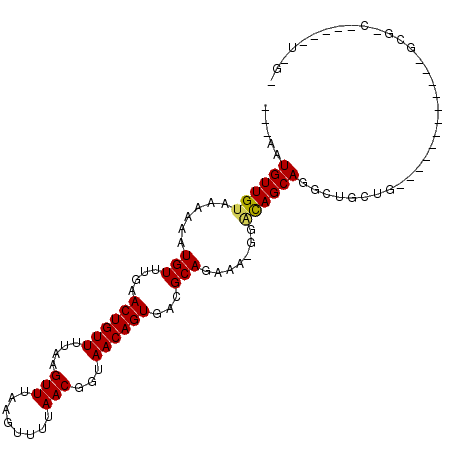
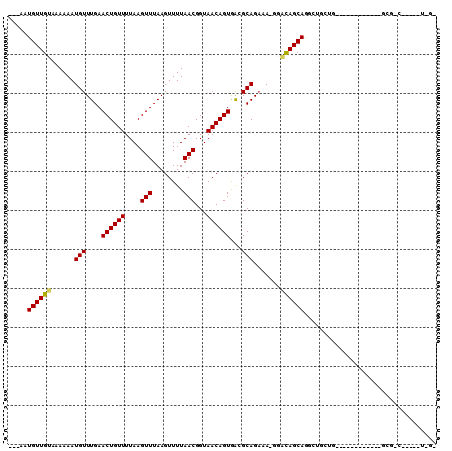
>2L_DroMel_CAF1 2196967 92 + 22407834 ---UGUGUUGUAAAAAAUGUUUGAACUGUUUUAAGUUUAAGUUUUAACGGUAACAGUAGAGCAGCAA-GUGCAGCAGGUCGAUCU---------AUGCGCC----GU-AA ---..(((((((.....(((((..((((((....(((........)))...)))))).)))))....-.)))))))((.((....---------...))))----..-.. ( -22.40) >DroVir_CAF1 21168 84 + 1 GGGGAUGUUGUAAAAAAUGUUUGAACUGUUUUAAGUUUAAGUUUUAACGGUAACAGUGACGCAGAAAUGAACAGCAGGCUGCUG-------------------------- (.((.((((((......(((....((((((....(((........)))...))))))...))).......))))))..)).)..-------------------------- ( -16.82) >DroPse_CAF1 40244 103 + 1 ---AAUGUUGUAAAAAAUGUUUGAACUGUUUUAAGUUUAAGUUUUAACGGUAACAGUGACGCAGAAACGGACAGCAGGCUGCUUCCACUCUUGAACGCGGC----AUUGC ---..((((((......(((....((((((....(((........)))...))))))...))).......)))))).((((((((.......))).)))))----..... ( -23.92) >DroGri_CAF1 9675 82 + 1 --GCAUGUUGUAAAAAAUGUUUGAACUGUUUUAAGUUUAAGUUUUAACGGUAACAGUGACGCAGAAAUGGGCAGCAGGUUGCUG-------------------------- --(((((((((......(((....((((((....(((........)))...))))))...))).......))))))...)))..-------------------------- ( -17.62) >DroAna_CAF1 6371 92 + 1 ---AAUGUUGUAAAAAAUGUUUGAACUGUUUUAAGUUUAAGUUUUAACGGUAACAGUAGAGCAGAAA-GAUUAGCAAGUAAAC-------------GGGCAACAUGU-GG ---.(((((((......((((((.((((((....(((........)))...))))))...((.....-.....))...)))))-------------).)))))))..-.. ( -16.80) >DroPer_CAF1 41602 103 + 1 ---AAUGUUGUAAAAAAUGUUUGAACUGUUUUAAGUUUAAGUUUUAACGGUAACAGUGACGCAGAAACGGACAGCAGGCUGCUUCCACUCUUGAACGCGGC----AUUGC ---..((((((......(((....((((((....(((........)))...))))))...))).......)))))).((((((((.......))).)))))----..... ( -23.92) >consensus ___AAUGUUGUAAAAAAUGUUUGAACUGUUUUAAGUUUAAGUUUUAACGGUAACAGUGACGCAGAAA_GGACAGCAGGCUGCUG____________GCG_C_____U_G_ .....((((((......(((....((((((....(((........)))...))))))...))).......)))))).................................. (-14.14 = -14.00 + -0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:10 2006