| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,798,867 – 21,798,981 |
| Length | 114 |
| Max. P | 0.937120 |

| Location | 21,798,867 – 21,798,981 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 78.26 |
| Mean single sequence MFE | -23.33 |
| Consensus MFE | -14.58 |
| Energy contribution | -15.70 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937120 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21798867 114 + 22407834 GUAAAUGGUUCUCUGCUUGGAAAUACCAUCAUCUGUUGGUAUUUUUAUGAACUUUUAAAUCAAA-UGCUCAAUUCGGUUUUAAAAGUUCUGGUUUCAAAAGUUCCCAAGAUUAAA ...............(((((((((((((........))))))))....(((((((((((((.((-(.....))).)).)))))))))))...............)))))...... ( -24.20) >DroSec_CAF1 41536 92 + 1 -----------------------UACCGACAUCUGUUGGUAUUUUUAUAAACUUUUAAUUCAAAGUGCUAAGUUCGCUUUUAAAAGUUCUGAUUUCAGAAGUUCCCAAGAUCAAA -----------------------(((((((....))))))).........((((((((....(((((.......)))))))))))))((((....))))................ ( -17.60) >DroSim_CAF1 34233 115 + 1 AUAAAUUGAUCUCUGCUUGGAAAUACCGACAUCUGUUGGUGUUUUAAUAAACUUUUAAUUCAAAGUGCUAAGUUCGCUUUUAAAAGUUCUGAUUUCAAAAGUUCCCAAGAUCAAA .....(((((((..((((.(((((((((((....)))))))))))....(((((((((....(((((.......))))))))))))))..........)))).....))))))). ( -28.20) >consensus _UAAAU_G_UCUCUGCUUGGAAAUACCGACAUCUGUUGGUAUUUUUAUAAACUUUUAAUUCAAAGUGCUAAGUUCGCUUUUAAAAGUUCUGAUUUCAAAAGUUCCCAAGAUCAAA ...................(((((((((((....)))))))))))....(((((((((....(((((.......)))))))))))))).((((((............)))))).. (-14.58 = -15.70 + 1.12)

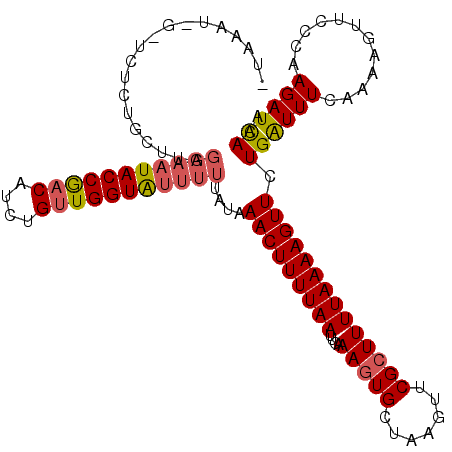
| Location | 21,798,867 – 21,798,981 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 78.26 |
| Mean single sequence MFE | -21.43 |
| Consensus MFE | -12.65 |
| Energy contribution | -12.43 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.59 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.525657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21798867 114 - 22407834 UUUAAUCUUGGGAACUUUUGAAACCAGAACUUUUAAAACCGAAUUGAGCA-UUUGAUUUAAAAGUUCAUAAAAAUACCAACAGAUGAUGGUAUUUCCAAGCAGAGAACCAUUUAC (((...((((((((............(((((((((((..(((((.....)-)))).))))))))))).......(((((.(....).)))))))))))))...)))......... ( -24.30) >DroSec_CAF1 41536 92 - 1 UUUGAUCUUGGGAACUUCUGAAAUCAGAACUUUUAAAAGCGAACUUAGCACUUUGAAUUAAAAGUUUAUAAAAAUACCAACAGAUGUCGGUA----------------------- ..((((.(..((....))..).))))((((((((((...((((........))))..)))))))))).......((((.((....)).))))----------------------- ( -16.20) >DroSim_CAF1 34233 115 - 1 UUUGAUCUUGGGAACUUUUGAAAUCAGAACUUUUAAAAGCGAACUUAGCACUUUGAAUUAAAAGUUUAUUAAAACACCAACAGAUGUCGGUAUUUCCAAGCAGAGAUCAAUUUAU .((((((((.((((............((((((((((...((((........))))..))))))))))........(((.((....)).)))..)))).....))))))))..... ( -23.80) >consensus UUUGAUCUUGGGAACUUUUGAAAUCAGAACUUUUAAAAGCGAACUUAGCACUUUGAAUUAAAAGUUUAUAAAAAUACCAACAGAUGUCGGUAUUUCCAAGCAGAGA_C_AUUUA_ ....(((((((.......((....))((((((((((...((((........))))..)))))))))).........)))..)))).............................. (-12.65 = -12.43 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:43:13 2006