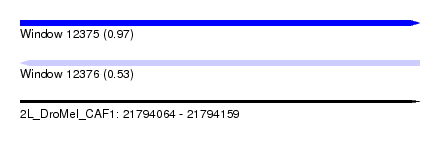
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,794,064 – 21,794,159 |
| Length | 95 |
| Max. P | 0.967144 |

| Location | 21,794,064 – 21,794,159 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 76.30 |
| Mean single sequence MFE | -24.99 |
| Consensus MFE | -14.05 |
| Energy contribution | -15.28 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
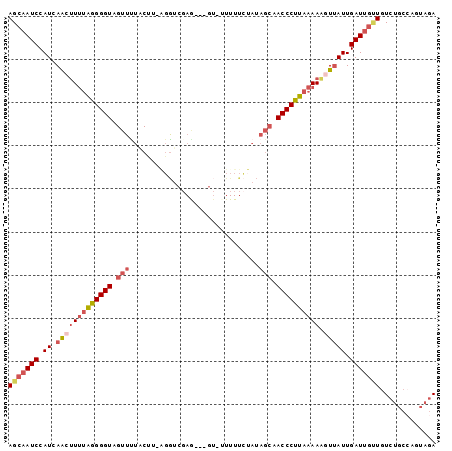
>2L_DroMel_CAF1 21794064 95 + 22407834 AGCAAUCCAUCAACUUUUGGGGGUUGUUUUACCCGAGGUCGAG---GU-UUUUUCUAUAGCAACCCUUAAAAAGUUAUUGAUUGUUGUCUGCCAGUAGA (((((((.((.(((((((((((((((((..(((((....)).)---))-.........)))))))))).))))))))).))))))).((((....)))) ( -29.30) >DroSec_CAF1 37410 94 + 1 AGCAAUCCAUCAGCUUUUGGGGGUUGUUUUACUG-GGGUCGAA---GU-UUUUUCUAUAGCAACCCUUAAAAAGUUAUUGAUUGUUGUCUGCCAGUAGA (((((((.((.(((((((((((((((((....((-(((.....---..-...))))).)))))))))).))))))))).))))))).((((....)))) ( -26.70) >DroSim_CAF1 29792 94 + 1 AGCAAUCCAUCAGCUUUUGGGGGUUGUUUUACUU-GGGUCGAG---GU-UUUUUCUAUAGCAACCCUUAAAAAGUUAUUGAUUGUUGUCUGCCAGUAGA (((((((.((.(((((((((((((((((.....(-(((.....---..-....)))).)))))))))).))))))))).))))))).((((....)))) ( -26.10) >DroEre_CAF1 40778 83 + 1 AGCAAUCCAUCAAAUUUUAGGGGUAGUUUUUCUU-AUGU---------------AUAUAGCAACCCCUAAAAAGUUAUUGAUUGAUGUGUGCGGGUAGA .(((...((((((.((((((((((.(((......-....---------------....))).)))))))))).........))))))..)))....... ( -21.34) >DroYak_CAF1 30418 94 + 1 AGCAAUCCAUCAAAAUUAAGGGGUAGUUCUCCUU-AGGUCGAG---UU-AUUUUCUAUAGCAACCCUUAAAAUGUUAUUGAUUGCUGUCUGCCAGUAGA (((((((.........(((((((.....))))))-)(((...(---((-((.....))))).)))..............))))))).((((....)))) ( -20.90) >DroPer_CAF1 52446 90 + 1 AGCAAUCCAUCAACUUCUAGGGGUAGUUUUUCCUUCGAACCAGGGUGAAAUUUU-------CACCCUUAAAAUGUGAUUGAUCUCUGUCGA--AAGAGA ..((((((((....(((.(((((.......))))).)))..(((((((.....)-------))))))....))).))))).(((((.....--.))))) ( -25.60) >consensus AGCAAUCCAUCAACUUUUAGGGGUAGUUUUACUU_AGGUCGAG___GU_UUUUUCUAUAGCAACCCUUAAAAAGUUAUUGAUUGUUGUCUGCCAGUAGA (((((((.((.(((((((((((((.(((..............................))).)))))))))).))))).)))))))............. (-14.05 = -15.28 + 1.22)


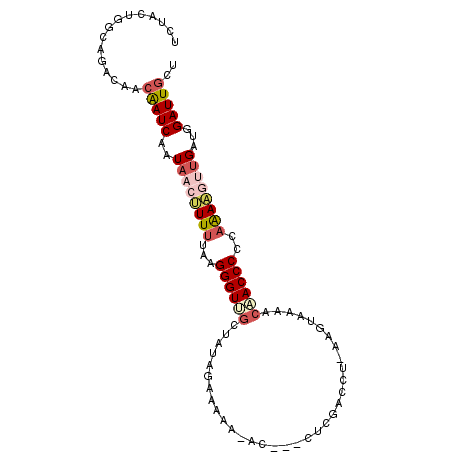
| Location | 21,794,064 – 21,794,159 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 76.30 |
| Mean single sequence MFE | -18.32 |
| Consensus MFE | -10.10 |
| Energy contribution | -11.26 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.55 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.527846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21794064 95 - 22407834 UCUACUGGCAGACAACAAUCAAUAACUUUUUAAGGGUUGCUAUAGAAAAA-AC---CUCGACCUCGGGUAAAACAACCCCCAAAAGUUGAUGGAUUGCU ...............(((((..((((((((...(((((((....).....-((---(.((....)))))....))))))..))))))))...))))).. ( -21.30) >DroSec_CAF1 37410 94 - 1 UCUACUGGCAGACAACAAUCAAUAACUUUUUAAGGGUUGCUAUAGAAAAA-AC---UUCGACCC-CAGUAAAACAACCCCCAAAAGCUGAUGGAUUGCU ......(((((.(....((((....(((((...((((((.....(((...-..---))).((..-..))....))))))..))))).)))).).))))) ( -14.80) >DroSim_CAF1 29792 94 - 1 UCUACUGGCAGACAACAAUCAAUAACUUUUUAAGGGUUGCUAUAGAAAAA-AC---CUCGACCC-AAGUAAAACAACCCCCAAAAGCUGAUGGAUUGCU ......(((((.(....((((....(((((...((((((...........-..---........-........))))))..))))).)))).).))))) ( -13.65) >DroEre_CAF1 40778 83 - 1 UCUACCCGCACACAUCAAUCAAUAACUUUUUAGGGGUUGCUAUAU---------------ACAU-AAGAAAAACUACCCCUAAAAUUUGAUGGAUUGCU .......(((..((((((.........((((((((((........---------------....-..........)))))))))).))))))...))). ( -19.85) >DroYak_CAF1 30418 94 - 1 UCUACUGGCAGACAGCAAUCAAUAACAUUUUAAGGGUUGCUAUAGAAAAU-AA---CUCGACCU-AAGGAGAACUACCCCUUAAUUUUGAUGGAUUGCU .............(((((((.....((..(((((((....(((.....))-).---(((.....-...)))......)))))))...))...))))))) ( -17.30) >DroPer_CAF1 52446 90 - 1 UCUCUU--UCGACAGAGAUCAAUCACAUUUUAAGGGUG-------AAAAUUUCACCCUGGUUCGAAGGAAAAACUACCCCUAGAAGUUGAUGGAUUGCU (((((.--.....))))).(((((.((((...((((((-------(.....)))))))..(((..(((..........))).)))...))))))))).. ( -23.00) >consensus UCUACUGGCAGACAACAAUCAAUAACUUUUUAAGGGUUGCUAUAGAAAAA_AC___CUCGACCU_AAGUAAAACAACCCCCAAAAGUUGAUGGAUUGCU ...............(((((..((((((((...((((((..................................))))))..))))))))...))))).. (-10.10 = -11.26 + 1.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:43:11 2006