| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,788,758 – 21,788,867 |
| Length | 109 |
| Max. P | 0.918010 |

| Location | 21,788,758 – 21,788,867 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.89 |
| Mean single sequence MFE | -22.02 |
| Consensus MFE | -12.60 |
| Energy contribution | -12.38 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918010 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
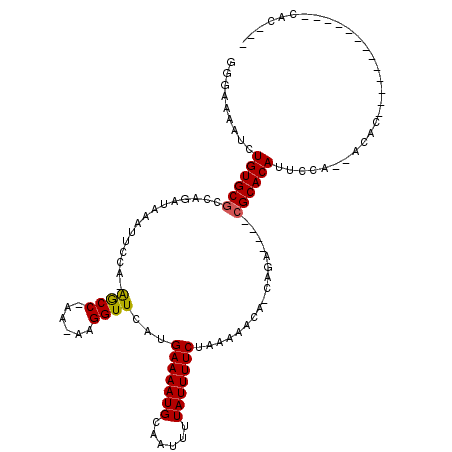
>2L_DroMel_CAF1 21788758 109 - 22407834 GUGA-AAUCUGUGCGCCAGAUAAAUUCCA-AGCCCAA-AAGGUUCAUGAAAAUGCAAUUUUAUUUUCUAAAAACA-UAGA----CGCACAUUCCAGCGCACUCGGACGGCCGAACAC--- (((.-.....((((((..((.........-((((...-..))))...(((((((......)))))))........-....----.......))..))))))((((....)))).)))--- ( -22.10) >DroVir_CAF1 38076 92 - 1 GGGAAAAUCUGUGCACCAGAUAAAUUCCU-GGCC-AA--AGGUUCAUGAAAAUGCAAUUUUAUUUUCUAAAAACA-CAGA----CGCACAUUCC---ACAC-------------CAC--- .((((....(((((.((((........))-))..-..--..(((...(((((((......)))))))....))).-....----.)))))))))---....-------------...--- ( -19.30) >DroPse_CAF1 43376 115 - 1 CUGAAAAUCUGUGCGCCAGAUAAAUUCCACAACCCAAAAGGGUUCAUGAAAAUGCAAUUUUAUUUUCUAAAAACA-CAGAGACACGCACAUUCCAACGCAUGU----GACUAUACAGACA (((....((((((.................(((((....)))))...(((((((......)))))))......))-))))......(((((........))))----)......)))... ( -22.10) >DroGri_CAF1 26667 96 - 1 CGGAAAAUCUGUGCGCCAGAUAAAUUCCG-GGCC-AA-AAGGUUCAUGAAAAUGCAAUUUUAUUUUCUAAAAACA-CAAA----CGCACAUUCCACCACAC-------------CAC--- .((((....((((((.............(-((((-..-..)))))..(((((((......)))))))........-....----)))))))))).......-------------...--- ( -21.40) >DroMoj_CAF1 35530 93 - 1 GGGAAAAUCUGUGCGCCAGAUAAAUUCCU-GGCC-AA--AGGUUCAUGAAAAUGCAAUUUUAUUUUCUAAAAACAGCAGA----CGCACAGAUU---CCAC-------------CAC--- ((...((((((((((((((........))-))).-..--..(((...(((((((......)))))))....)))......----.)))))))))---...)-------------)..--- ( -29.10) >DroAna_CAF1 27622 91 - 1 GGGA-AAUCUGUGCGCCAGAUAAAUUCCA-AGCCCAA-AAGGUUCAUGAAAAUGCAAUUUUAUUUUCUAAAAACA-CAGA----CGCACAUUCCAG------------------CAC--- .(((-(...((((((.(............-((((...-..))))...(((((((......)))))))........-..).----))))))))))..------------------...--- ( -18.10) >consensus GGGAAAAUCUGUGCGCCAGAUAAAUUCCA_AGCC_AA_AAGGUUCAUGAAAAUGCAAUUUUAUUUUCUAAAAACA_CAGA____CGCACAUUCCA__ACAC_____________CAC___ .........((((((...............((((......))))...(((((((......))))))).................)))))).............................. (-12.60 = -12.38 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:43:10 2006