| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,777,998 – 21,778,094 |
| Length | 96 |
| Max. P | 0.751619 |

| Location | 21,777,998 – 21,778,094 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.09 |
| Mean single sequence MFE | -29.42 |
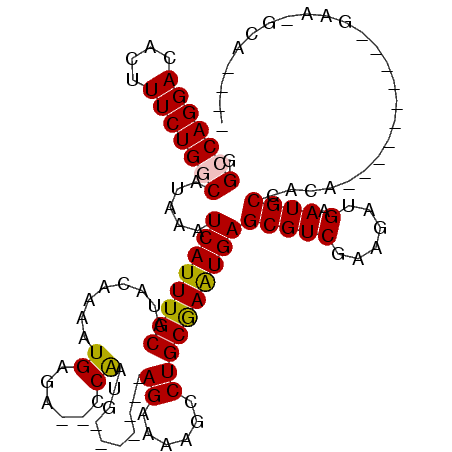
| Consensus MFE | -18.08 |
| Energy contribution | -17.83 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21777998 96 + 22407834 GGCCAGGACACUUUCUGGCAUAAAUCAUUUGCAUUCAAAAUGAGA--CCAAUG-------AGGAAAGCCUGCGAAUGAGCGUCGAAGAUGAAUGCCGCA----------GAA-ACA---- .(((((((....)))))))...........(((((((...(..((--(.....-------(((....)))((......)))))..)..)))))))....----------...-...---- ( -24.90) >DroPse_CAF1 31531 120 + 1 UGGCAGGACACUUUCUGGCAUAAAUCAUUUGCAUACGAAAUGAGAGGCCGAUGCGAGAGCAGAGAGGCCUGCGAGUGAGCGUCGAAGAUGAAUGCGAGAACAACACCCAGAAGGCAGUGG ........((((((((((......((((((((...(.......)(((((..(((....)))....)))))))))))))(((((......).))))...........))))))...)))). ( -36.80) >DroEre_CAF1 22701 96 + 1 GGCCAGGACACUUUCUGGCAUAAAUCAUUUGCAUUCAAAAUGAGA--CCAAUG-------AGGAAAGCCUGCAAAUGAGCGUCGAAGAUGAAUGCCGCA----------GAA-GCC---- .(((((((....))))))).....((((((((.(((.....))).--......-------(((....)))))))))))(((.((........)).))).----------...-...---- ( -25.00) >DroYak_CAF1 9693 93 + 1 GGCCAGGACACUUUCUGGCAUAAAUCAUUUGCAUUCAAAAUGAGA--CCAAUG-------AGGAAAGCCUGCGAAUGAGCGUCGAAGAUGAAUGCCGGA----------GAG-------- ..........(((((((((((...((((((((.(((.....))).--......-------(((....))))))))))).((((...)))).))))))))----------)))-------- ( -28.80) >DroAna_CAF1 17649 102 + 1 GGCCAGGACACUUUCUGGCAUAAAUCAUUUGCAUACAAAAUGAGA--CCAAUG-------AGAGGUGCCUGCGAAUGAGCGUCGAAGAUGAAUGCGACAACAAUA----AAA-ACA---- .(((((((....))))))).....(((((((((..(.....)..(--((....-------...)))...)))))))))..((((..........)))).......----...-...---- ( -24.20) >DroPer_CAF1 33260 120 + 1 UGGCAGGACACUUUCUGGCAUAAAUCAUUUGCAUACGAAAUGAGAGGCCGAUGCGAGAGCAGAGAGGCCUGCGAGUGAGCGUCGAAGAUGAAUGCGAGAACAACACCCAGAAGGCAGUGG ........((((((((((......((((((((...(.......)(((((..(((....)))....)))))))))))))(((((......).))))...........))))))...)))). ( -36.80) >consensus GGCCAGGACACUUUCUGGCAUAAAUCAUUUGCAUACAAAAUGAGA__CCAAUG_______AGAAAAGCCUGCGAAUGAGCGUCGAAGAUGAAUGCCACA__________GAA_GCA____ .(((((((....))))))).....((((((((........((......))..........((......))))))))))(((((......).))))......................... (-18.08 = -17.83 + -0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:43:08 2006