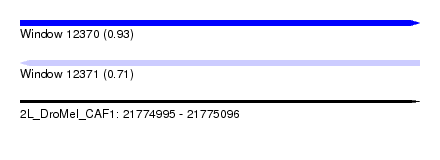
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,774,995 – 21,775,096 |
| Length | 101 |
| Max. P | 0.934558 |

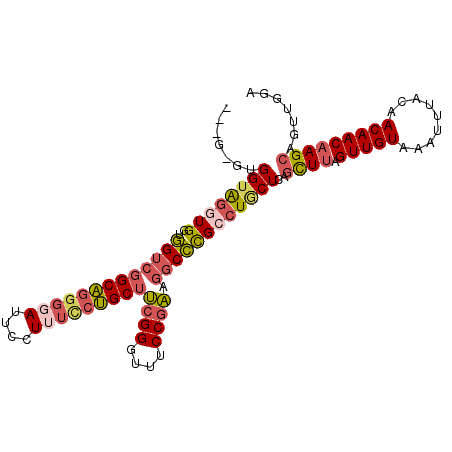
| Location | 21,774,995 – 21,775,096 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 81.38 |
| Mean single sequence MFE | -38.92 |
| Consensus MFE | -29.21 |
| Energy contribution | -30.23 |
| Covariance contribution | 1.02 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21774995 101 + 22407834 ---G-GUGGUAGGUGGUGGUCGGCAGGGUAUUCCUUUCCUGCUUCGGGUUUCCGAUGGCCCGCCUGCUCAGCUUAGUUGUAAAUUUACAACAACAAGCAGUUGGA ---.-.((((((((((..((((((((((........)))))))((((....)))).))))))))))).))((((.((((((....))))))...))))....... ( -38.70) >DroSec_CAF1 16742 101 + 1 ---G-GUGGUAGGUGGUGGUCGGCAGGGGAUUCCUUUCCUGCUUCGGGUUUCCGAAGGCCCGCCUACUCAGCUUAGUUGUAAAUUUAGAACAACAAGCAAUUGGA ---.-.((((((((((..(((.(((((..(....)..)))))(((((....)))))))))))))))).))((((.(((((.........)))))))))....... ( -40.10) >DroSim_CAF1 9519 101 + 1 ---G-GUGGUAGGUGGUGGUCGGCAGGGGAUUCCUUUCCUGCUUCGGGUUUCCGAAGGCCCGCCUACUCAGCUUAGUUGUAAAUUUAGAACAACAAGCAAUUGGA ---.-.((((((((((..(((.(((((..(....)..)))))(((((....)))))))))))))))).))((((.(((((.........)))))))))....... ( -40.10) >DroEre_CAF1 19636 101 + 1 ---G-GUGGUAGGUGGUGGUCGGCAGGGGAUUCCUUUCCUGCUUCGGAUUUCCGACGGCCCGACUGCUCAGCUUAGUUGUAAAUUUACAACAACAAGCAGUUGGA ---.-............((((((((((..(....)..))))).((((....)))))))))((((((((.......((((((....))))))....)))))))).. ( -37.60) >DroYak_CAF1 6883 104 + 1 UAGG-GUGGUAGGUGAUGGUCGGCAGGGGAUUCCUUUUCUGCUUCGGUUUUCCGACGGCCCGCCUGCUCAGCUUAGUUGUAAAUUUACAACAACAAGCAGUUAGA ((((-.(((((((((..(((((((((..((....))..)))).((((....)))))))))))))))).)).))))((((((....)))))).............. ( -41.60) >DroAna_CAF1 15607 95 + 1 UGGAGGGGGGGAGUGGGAGUGGGCUUGGG----------GGCGAAGGAUUCCCAUAGCCUUGACUGCUCAGUUCAGUUGUGAAUUUACAACAACCAGAAGCCGGA .((....(((((.((((((((((((((((----------(((....).)))))).)))))..))).)))).))).((((((....))))))..)).....))... ( -35.40) >consensus ___G_GUGGUAGGUGGUGGUCGGCAGGGGAUUCCUUUCCUGCUUCGGGUUUCCGAAGGCCCGCCUGCUCAGCUUAGUUGUAAAUUUACAACAACAAGCAGUUGGA .......((((((((..(((((((((((((....)))))))))((((....)))).))))))))))))..((((.(((((.........)))))))))....... (-29.21 = -30.23 + 1.02)

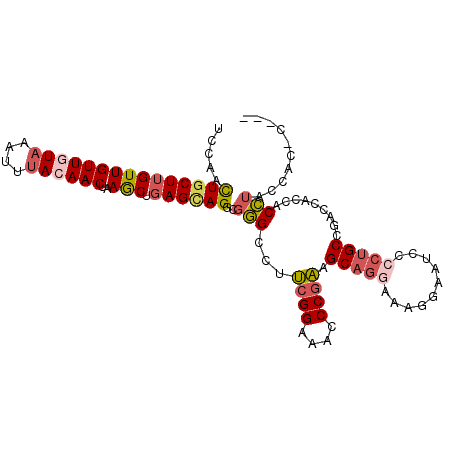
| Location | 21,774,995 – 21,775,096 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 81.38 |
| Mean single sequence MFE | -28.17 |
| Consensus MFE | -17.76 |
| Energy contribution | -18.52 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21774995 101 - 22407834 UCCAACUGCUUGUUGUUGUAAAUUUACAACUAAGCUGAGCAGGCGGGCCAUCGGAAACCCGAAGCAGGAAAGGAAUACCCUGCCGACCACCACCUACCAC-C--- (((..(((((((((((((((....))))))..))).))))))..)))...((((....)))).(((((..........))))).................-.--- ( -28.00) >DroSec_CAF1 16742 101 - 1 UCCAAUUGCUUGUUGUUCUAAAUUUACAACUAAGCUGAGUAGGCGGGCCUUCGGAAACCCGAAGCAGGAAAGGAAUCCCCUGCCGACCACCACCUACCAC-C--- .......(((((((((.........))))).))))((.(((((.((((.(((((....))))))).((..(((.....))).)).....)).))))))).-.--- ( -31.50) >DroSim_CAF1 9519 101 - 1 UCCAAUUGCUUGUUGUUCUAAAUUUACAACUAAGCUGAGUAGGCGGGCCUUCGGAAACCCGAAGCAGGAAAGGAAUCCCCUGCCGACCACCACCUACCAC-C--- .......(((((((((.........))))).))))((.(((((.((((.(((((....))))))).((..(((.....))).)).....)).))))))).-.--- ( -31.50) >DroEre_CAF1 19636 101 - 1 UCCAACUGCUUGUUGUUGUAAAUUUACAACUAAGCUGAGCAGUCGGGCCGUCGGAAAUCCGAAGCAGGAAAGGAAUCCCCUGCCGACCACCACCUACCAC-C--- (((.((((((((((((((((....))))))..))).))))))).)))..(((((...(((......))).(((.....))).))))).............-.--- ( -30.70) >DroYak_CAF1 6883 104 - 1 UCUAACUGCUUGUUGUUGUAAAUUUACAACUAAGCUGAGCAGGCGGGCCGUCGGAAAACCGAAGCAGAAAAGGAAUCCCCUGCCGACCAUCACCUACCAC-CCUA .....(((((((((((((((....))))))..))).))))))..(((...((((....)))).((((....((....))))))................)-)).. ( -26.30) >DroAna_CAF1 15607 95 - 1 UCCGGCUUCUGGUUGUUGUAAAUUCACAACUGAACUGAGCAGUCAAGGCUAUGGGAAUCCUUCGCC----------CCCAAGCCCACUCCCACUCCCCCCCUCCA ...((((.(((((((((((......)))))..)))).)).))))..((((.((((...........----------))))))))..................... ( -21.00) >consensus UCCAACUGCUUGUUGUUGUAAAUUUACAACUAAGCUGAGCAGGCGGGCCUUCGGAAACCCGAAGCAGGAAAGGAAUCCCCUGCCGACCACCACCUACCAC_C___ .....(((((((((((((((....))))))..))).))))))..(((...((((....)))).(((((..........))))).........))).......... (-17.76 = -18.52 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:43:07 2006