
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,770,033 – 21,770,153 |
| Length | 120 |
| Max. P | 0.812160 |

| Location | 21,770,033 – 21,770,153 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.20 |
| Mean single sequence MFE | -40.98 |
| Consensus MFE | -30.17 |
| Energy contribution | -27.15 |
| Covariance contribution | -3.02 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.776679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
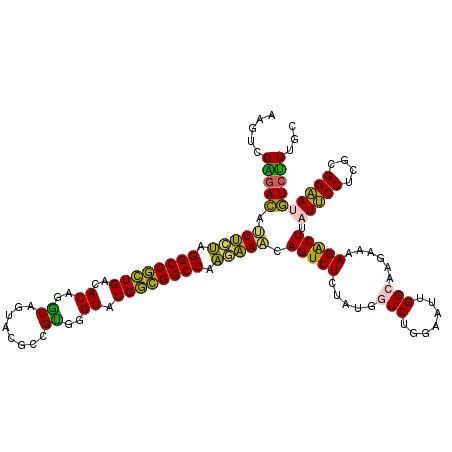
>2L_DroMel_CAF1 21770033 120 + 22407834 AAGUCGAGACAUUUCUAGGCUGCUGACACAGGCAGUUCGCCGUGGGUAUGGCGGCCAAGAAACAGUCUCUAUGGUCUGGAAUGGGCAAGAAAAGACUUAUUCCUCGCGGAAUGUCUUUCC ((((((((((.(((((.((((((((.(((.(((.....))))))....)))))))).)))))..)))))....((((.....)))).......)))))(((((....)))))........ ( -42.80) >DroPse_CAF1 22821 118 + 1 AA--AGGGAUACUCGAGGGCUUGUGGCACAGGCAGUACUCUGCGGGUAUGUGGGCCAUCGAGCAGUCUCGCUGUUCGGGUAUUGGCAGGAACAGACUUAUCCCUCUUGGGAGAUCCUCUU ..--.(((((.(((((.((((..((..((..((((....))))..)).))..)))).)))))..))))).((((((..((....))..)))))).....((((....))))......... ( -43.70) >DroSec_CAF1 12000 120 + 1 AAGUCGAGACAUUUCUAGGCUGCUGACACAGGCAGUUCGCCGUGGGUAUGGCGGCCAAGAAACAGUCUCUAUGGUCUGGAAUUGGCAAGAAAAGACUUAUUCCCCGCGGAAUGUCUUUGC ...(((((((.(((((.((((((((.(((.(((.....))))))....)))))))).)))))..)))))....(((.......)))..))((((((..(((((....))))))))))).. ( -41.60) >DroSim_CAF1 4778 120 + 1 AAGUCGAGACAUUUCUAGGCUGCUGACACAGGCAGUUCGCCGUGGGUAUGGCGGCCAAGAAACAGUCUCUAUGGUCUGGAAUUGGCAAGAAAAGACUUAUUCCCCGCGGAAUGUCUUUGC ...(((((((.(((((.((((((((.(((.(((.....))))))....)))))))).)))))..)))))....(((.......)))..))((((((..(((((....))))))))))).. ( -41.60) >DroEre_CAF1 14335 120 + 1 AAGUUGGAACAUUUGUAGGCUGCUGACACAGACAGUAAGCGGUGGGUAUGGCGGCCAAGAAACAGUCUCUAUGGUCCGGAAUAGGCAAGAAAAGGCUUAUUCCUCGCGGAAUGUCUUUGC ..((.(((.(((((....((((((.((.......)).)))))).......((((((((((.......))).))))).(((((((((........)))))))))..)).))))))))..)) ( -34.10) >DroPer_CAF1 23623 118 + 1 AA--AGGGAUACUCGAGGGCUUGUGGCACAUGCAGUACUCUGCGGGUAUGUGGGCCAUCGAACAGUCUCGCUGUUCGGGUAUUGGCAGGAACAGACUUAUCCCUCUUGGGAGAUCCUCUU ..--((((((((((((.((((..((..((.(((((....))))).)).))..)))).))((((((.....)))))))))))))................((((....))))...)))... ( -42.10) >consensus AAGUCGAGACAUUUCUAGGCUGCUGACACAGGCAGUACGCCGUGGGUAUGGCGGCCAAGAAACAGUCUCUAUGGUCUGGAAUUGGCAAGAAAAGACUUAUUCCUCGCGGAAUGUCUUUGC .....(((((.(((((.((((((((..((..((........))..)).)))))))).))))).(((((.....(((.......)))......))))).(((((....))))))))))... (-30.17 = -27.15 + -3.02)

| Location | 21,770,033 – 21,770,153 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.20 |
| Mean single sequence MFE | -35.32 |
| Consensus MFE | -25.29 |
| Energy contribution | -24.47 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812160 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
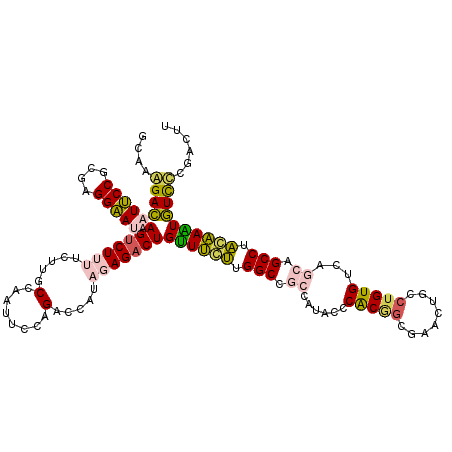
>2L_DroMel_CAF1 21770033 120 - 22407834 GGAAAGACAUUCCGCGAGGAAUAAGUCUUUUCUUGCCCAUUCCAGACCAUAGAGACUGUUUCUUGGCCGCCAUACCCACGGCGAACUGCCUGUGUCAGCAGCCUAGAAAUGUCUCGACUU (((((((((((((....)))))..))))))))...................(((((.((((((.(((.((...((.((.(((.....))))).))..)).))).)))))))))))..... ( -42.20) >DroPse_CAF1 22821 118 - 1 AAGAGGAUCUCCCAAGAGGGAUAAGUCUGUUCCUGCCAAUACCCGAACAGCGAGACUGCUCGAUGGCCCACAUACCCGCAGAGUACUGCCUGUGCCACAAGCCCUCGAGUAUCCCU--UU ..(.((....))).((((((((..(.((((((............)))))))......((((((.(((.....(((..((((....))))..)))......))).))))))))))))--)) ( -35.10) >DroSec_CAF1 12000 120 - 1 GCAAAGACAUUCCGCGGGGAAUAAGUCUUUUCUUGCCAAUUCCAGACCAUAGAGACUGUUUCUUGGCCGCCAUACCCACGGCGAACUGCCUGUGUCAGCAGCCUAGAAAUGUCUCGACUU (((((((((((((....)))))..))))....))))...............(((((.((((((.(((.((...((.((.(((.....))))).))..)).))).)))))))))))..... ( -35.90) >DroSim_CAF1 4778 120 - 1 GCAAAGACAUUCCGCGGGGAAUAAGUCUUUUCUUGCCAAUUCCAGACCAUAGAGACUGUUUCUUGGCCGCCAUACCCACGGCGAACUGCCUGUGUCAGCAGCCUAGAAAUGUCUCGACUU (((((((((((((....)))))..))))....))))...............(((((.((((((.(((.((...((.((.(((.....))))).))..)).))).)))))))))))..... ( -35.90) >DroEre_CAF1 14335 120 - 1 GCAAAGACAUUCCGCGAGGAAUAAGCCUUUUCUUGCCUAUUCCGGACCAUAGAGACUGUUUCUUGGCCGCCAUACCCACCGCUUACUGUCUGUGUCAGCAGCCUACAAAUGUUCCAACUU (((.(((((....(((.((((((.((........)).))))))((.(((.(((((...))))))))))...........)))....))))).)))......................... ( -26.90) >DroPer_CAF1 23623 118 - 1 AAGAGGAUCUCCCAAGAGGGAUAAGUCUGUUCCUGCCAAUACCCGAACAGCGAGACUGUUCGAUGGCCCACAUACCCGCAGAGUACUGCAUGUGCCACAAGCCCUCGAGUAUCCCU--UU (((.(((((((....(((((..............((((.....(((((((.....))))))).)))).(((((....((((....))))))))).......)))))))).))))))--). ( -35.90) >consensus GCAAAGACAUUCCGCGAGGAAUAAGUCUUUUCUUGCCAAUUCCAGACCAUAGAGACUGUUUCUUGGCCGCCAUACCCACGGCGAACUGCCUGUGUCAGCAGCCUACAAAUGUCCCGACUU ....(((((((((....))))).(((((((.....(........).....)))))))((((((.(((.((......(((((........)))))...)).))).))))))))))...... (-25.29 = -24.47 + -0.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:43:03 2006