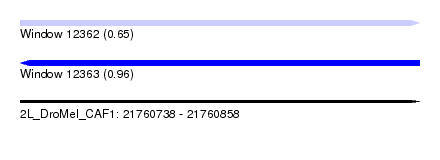
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,760,738 – 21,760,858 |
| Length | 120 |
| Max. P | 0.962646 |

| Location | 21,760,738 – 21,760,858 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.42 |
| Mean single sequence MFE | -34.92 |
| Consensus MFE | -21.02 |
| Energy contribution | -21.78 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21760738 120 + 22407834 AAAUUUCUGAAGGCACUCAGUGCAUUCAAAAAUUUUUUCAUUUGUAUGGUUCCUUAGGUGAGCCCUAAGCGAGUGCUUUUUGAAAUUCUUCGGGCACAAAGGGCACUGAAGAACCUCGUU ...((((.((((((((((.(((((..................))))).....((((((.....)))))).)))))))))).))))((((((((((.......)).))))))))....... ( -36.17) >DroGri_CAF1 14211 120 + 1 AAACUUGGCCGAGCACUCGACACACUCGAAGAUCCUUUCGUCCGUAUGAUUGCGCAGAUGGGGACGCAGUGAGUGCUUGCGAAACUGUUUGGGGCACAGCGGACACUGGAGCAGCUCAUU ..........((((..((((.....)))).(.(((....((((((.(.(((((((.......).)))))).)((((((.((((....)))))))))).))))))...))).).))))... ( -36.80) >DroSim_CAF1 13781 120 + 1 AAAUUUCUGAAGGCACUCCGUGCAUUCAAAUAUUUUUUCAUUUGUAUGGUUCCUUAGGUGAGCCCGAAGAGAGUGCUUUUUGAAAUUCUUCGGGCACAAAGGGCACUGAAGAACCUCGUU ...((((.((((((((((((((((..................)))))((((((....).)))))....).)))))))))).))))((((((((((.......)).))))))))....... ( -35.07) >DroWil_CAF1 36243 120 + 1 AAACUUUUGCAAACACUCGGGGCAUUGAAAUAUCUUCUCAUCCGUGUGAUUACGUAAAUGCGCUCUUAGGGAAUGUUUACGAAAUUUUUUACUGCAUAGUGGACAUUGAAGAAGCUCAUU .............(((.((((.....(((.....)))...)))).)))........((((.((((((....(((((((((..................))))))))).))).))).)))) ( -23.17) >DroYak_CAF1 15663 120 + 1 AAAUUUCUGAAGGCACUCCGUGCAUUCAAAAAUUUUUUCAUUUGUAUGGUUCCUUAGGUGAGGCCUAAGGGAGUGCUUUUUGAAAUUCUUCGGGCACAAGGGGCACUGAAGAAGCUCGUU ...((((.((((((((.(((((((..................)))))))(((((((((.....))))))))))))))))).))))((((((((((.......)).))))))))....... ( -43.37) >consensus AAAUUUCUGAAGGCACUCCGUGCAUUCAAAAAUUUUUUCAUUUGUAUGGUUCCUUAGGUGAGCCCUAAGGGAGUGCUUUUUGAAAUUCUUCGGGCACAAAGGGCACUGAAGAAGCUCGUU ....(((.((((((((((...................((((....))))...((((((.....)))))).)))))))))).))).((((((((((.......)).))))))))....... (-21.02 = -21.78 + 0.76)

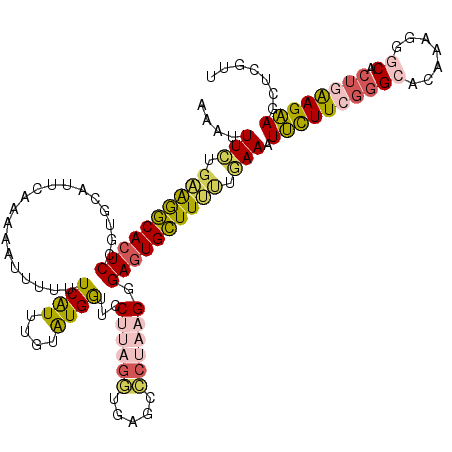
| Location | 21,760,738 – 21,760,858 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.42 |
| Mean single sequence MFE | -32.24 |
| Consensus MFE | -18.24 |
| Energy contribution | -19.04 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962646 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21760738 120 - 22407834 AACGAGGUUCUUCAGUGCCCUUUGUGCCCGAAGAAUUUCAAAAAGCACUCGCUUAGGGCUCACCUAAGGAACCAUACAAAUGAAAAAAUUUUUGAAUGCACUGAGUGCCUUCAGAAAUUU ...((((((((((.(..(.....)..)..)))))))))).........((.((((((.....))))))))...............(((((((((((.((((...)))).))))))))))) ( -35.50) >DroGri_CAF1 14211 120 - 1 AAUGAGCUGCUCCAGUGUCCGCUGUGCCCCAAACAGUUUCGCAAGCACUCACUGCGUCCCCAUCUGCGCAAUCAUACGGACGAAAGGAUCUUCGAGUGUGUCGAGUGCUCGGCCAAGUUU ...(((((...((.(((...(((((.......)))))..))).(((((((..(((((........)))))..(((((...((((......)))).)))))..))))))).))...))))) ( -36.60) >DroSim_CAF1 13781 120 - 1 AACGAGGUUCUUCAGUGCCCUUUGUGCCCGAAGAAUUUCAAAAAGCACUCUCUUCGGGCUCACCUAAGGAACCAUACAAAUGAAAAAAUAUUUGAAUGCACGGAGUGCCUUCAGAAAUUU ...(((((.((((.((((((((((.(((((((((................))))))))).))...)))).......((((((......))))))...)))))))).)))))......... ( -36.99) >DroWil_CAF1 36243 120 - 1 AAUGAGCUUCUUCAAUGUCCACUAUGCAGUAAAAAAUUUCGUAAACAUUCCCUAAGAGCGCAUUUACGUAAUCACACGGAUGAGAAGAUAUUUCAAUGCCCCGAGUGUUUGCAAAAGUUU ((((.(((.(((.(((((....((((.(((.....))).)))).)))))....)))))).))))...((((.(((.(((.(((((.....))))).....))).))).))))........ ( -19.10) >DroYak_CAF1 15663 120 - 1 AACGAGCUUCUUCAGUGCCCCUUGUGCCCGAAGAAUUUCAAAAAGCACUCCCUUAGGCCUCACCUAAGGAACCAUACAAAUGAAAAAAUUUUUGAAUGCACGGAGUGCCUUCAGAAAUUU ...(((.((((((.(..(.....)..)..)))))).)))...........(((((((.....)))))))................(((((((((((.((((...)))).))))))))))) ( -33.00) >consensus AACGAGCUUCUUCAGUGCCCUUUGUGCCCGAAGAAUUUCAAAAAGCACUCCCUUAGGGCUCACCUAAGGAACCAUACAAAUGAAAAAAUAUUUGAAUGCACCGAGUGCCUUCAGAAAUUU ...((((((((((.(.((.......))).)))))))))).....((((((.((((((.....))))))....(((.((((..........)))).)))....))))))............ (-18.24 = -19.04 + 0.80)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:43:00 2006