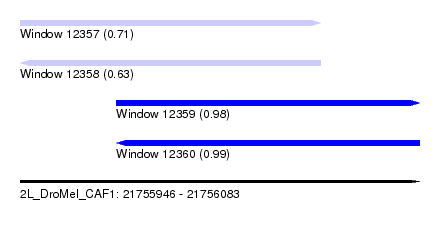
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,755,946 – 21,756,083 |
| Length | 137 |
| Max. P | 0.994453 |

| Location | 21,755,946 – 21,756,049 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 81.98 |
| Mean single sequence MFE | -33.58 |
| Consensus MFE | -20.78 |
| Energy contribution | -21.74 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
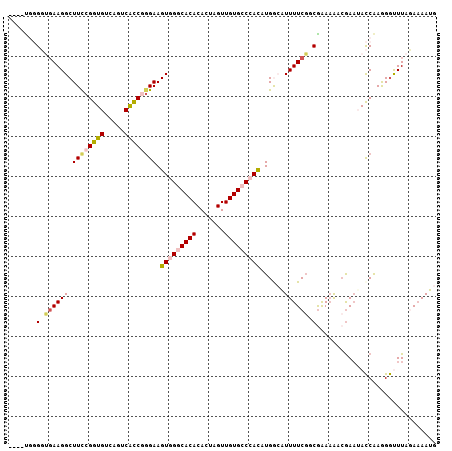
>2L_DroMel_CAF1 21755946 103 + 22407834 ----UGGGGUGAAGGCUUCCGGUGUCAGUCACCGAGAAGUGGGCACACACUAGUUGUGGCCACAUGGCAUUUUCGUCAAAAAACAAAUACCAAGGGUUUAGAAAAUG ----...((((...((((((((((.....))))).))))).(((.((((.....)))))))...((((......)))).........))))................ ( -29.40) >DroSec_CAF1 9529 103 + 1 ----UGGGGUGAAGGCUUCCGGUGUCAGUCACCGGGAAGUGGGCACACACUAGUUGUGCCCACAUGAAAUUUUCGGCGAAAAACGAAAACCAAGGGCUUAUAAAAUG ----........((((((((((((.....)))))))).(((((((((.......)))))))))......((((((........))))))......))))........ ( -36.70) >DroSim_CAF1 9538 102 + 1 ----UGGGGUGAAGGCUUCCGGUGUCAGUCACCGGGAAGUGGGCACACACUAGUUGUGCCCACAUGAAAUUUUCGGCGAA-AACGAAAACCAAGGGCUUAUAAAAUG ----........((((((((((((.....)))))))).(((((((((.......)))))))))......((((((.....-..))))))......))))........ ( -36.40) >DroEre_CAF1 8584 107 + 1 GCCCCCCGCAGAAGGCUUCCGGUGUCAGUCGUCAGGAAGUGGGCACACACUAGUUGUGGCGACAUGGCAUUUUCUACGGAAGACGAAUACCGAAGGUUUAGAAAAUG ((.....))...((((((.(((((((((((((((.((((((......))))..)).))))))).)))).(((((....))))).....)))).))))))........ ( -34.50) >DroYak_CAF1 11199 104 + 1 ---CCAUGCAUAAGGCUUUCGGUGUCAGUCACCAGGAAGUGGGCACACACUAGUUGUGCCGAUAUGGCGUUUUCGCCGAAAAAGGAUUAUCAAAGAUUUAGAUAAUG ---((.........((((((((((.....)))).)))))).((((((.......))))))....(((((....))))).....))((((((.........)))))). ( -30.90) >consensus ____UGGGGUGAAGGCUUCCGGUGUCAGUCACCGGGAAGUGGGCACACACUAGUUGUGCCCACAUGGCAUUUUCGGCGAAAAACGAAUACCAAGGGUUUAGAAAAUG .......(.((((((.((((((((.....)))))))).(((((((((.......)))))))))......)))))).).............................. (-20.78 = -21.74 + 0.96)

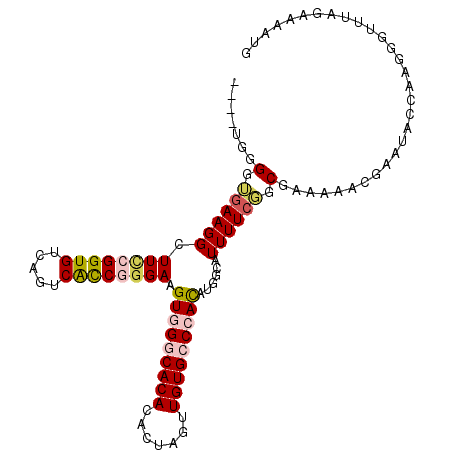
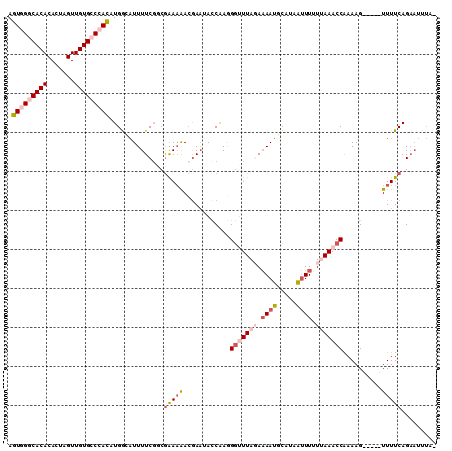
| Location | 21,755,946 – 21,756,049 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 81.98 |
| Mean single sequence MFE | -29.26 |
| Consensus MFE | -17.64 |
| Energy contribution | -19.08 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.625980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21755946 103 - 22407834 CAUUUUCUAAACCCUUGGUAUUUGUUUUUUGACGAAAAUGCCAUGUGGCCACAACUAGUGUGUGCCCACUUCUCGGUGACUGACACCGGAAGCCUUCACCCCA---- ...............(((((((((((....)))))..))))))((.(((((((.....)))).)))))((((.(((((.....)))))))))...........---- ( -28.40) >DroSec_CAF1 9529 103 - 1 CAUUUUAUAAGCCCUUGGUUUUCGUUUUUCGCCGAAAAUUUCAUGUGGGCACAACUAGUGUGUGCCCACUUCCCGGUGACUGACACCGGAAGCCUUCACCCCA---- ................((((((((((..(((((((.....))..(((((((((.(....)))))))))).....)))))..)))...))))))).........---- ( -32.80) >DroSim_CAF1 9538 102 - 1 CAUUUUAUAAGCCCUUGGUUUUCGUU-UUCGCCGAAAAUUUCAUGUGGGCACAACUAGUGUGUGCCCACUUCCCGGUGACUGACACCGGAAGCCUUCACCCCA---- ..........((....((((((((..-.....))))))))....(((((((((.(....))))))))))...((((((.....))))))..))..........---- ( -31.60) >DroEre_CAF1 8584 107 - 1 CAUUUUCUAAACCUUCGGUAUUCGUCUUCCGUAGAAAAUGCCAUGUCGCCACAACUAGUGUGUGCCCACUUCCUGACGACUGACACCGGAAGCCUUCUGCGGGGGGC ..........(((...)))....(((((((((((((....((.((((((((((.....)))).)).((.....))......))))..)).....))))))))))))) ( -28.60) >DroYak_CAF1 11199 104 - 1 CAUUAUCUAAAUCUUUGAUAAUCCUUUUUCGGCGAAAACGCCAUAUCGGCACAACUAGUGUGUGCCCACUUCCUGGUGACUGACACCGAAAGCCUUAUGCAUGG--- .((((((.........))))))....((((((((....)))).....((((((.(....)))))))........((((.....))))))))((.....))....--- ( -24.90) >consensus CAUUUUCUAAACCCUUGGUAUUCGUUUUUCGCCGAAAAUGCCAUGUGGGCACAACUAGUGUGUGCCCACUUCCCGGUGACUGACACCGGAAGCCUUCACCCCA____ ................(((.........................(((((((((.(....))))))))))...((((((.....))))))..)))............. (-17.64 = -19.08 + 1.44)

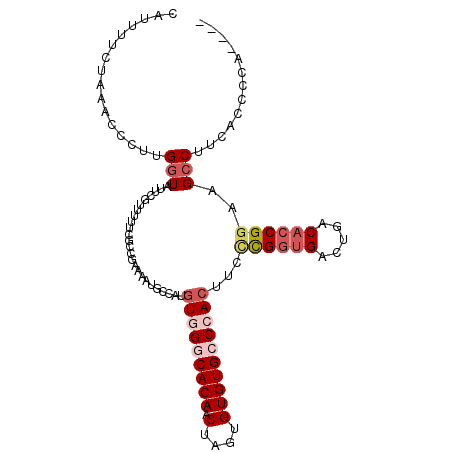
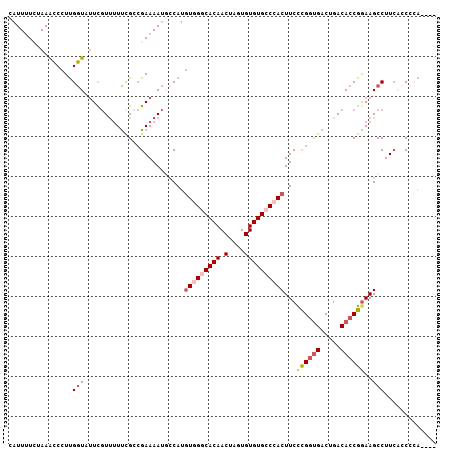
| Location | 21,755,979 – 21,756,083 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 79.80 |
| Mean single sequence MFE | -27.78 |
| Consensus MFE | -13.89 |
| Energy contribution | -16.17 |
| Covariance contribution | 2.28 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21755979 104 + 22407834 AGUGGGCACACACUAGUUGUGGCCACAUGGCAUUUUCGUCAAAAAACAAAUACCAAGGGUUUAGAAAAUGCAACAUUUCUUAAACCAAAAG-----UUUUCAGUUUUUA- .((((.((((.......)))).)))).((((......))))((((((....((....((((((..(((((...)))))..))))))....)-----).....)))))).- ( -26.50) >DroSec_CAF1 9562 104 + 1 AGUGGGCACACACUAGUUGUGCCCACAUGAAAUUUUCGGCGAAAAACGAAAACCAAGGGCUUAUAAAAUGCAUAAUUUUUUAAACCAAAAG-----UUUUCAGAAUUUA- .(((((((((.......))))))))).(((((((((((........)))))).....((.(((.(((((.....))))).))).)).....-----.))))).......- ( -28.30) >DroSim_CAF1 9571 103 + 1 AGUGGGCACACACUAGUUGUGCCCACAUGAAAUUUUCGGCGAA-AACGAAAACCAAGGGCUUAUAAAAUGCAUAAUUUUUUAAACCAAAAG-----UUUUCAGAAUUUA- .(((((((((.......))))))))).(((((((((((.....-..)))))).....((.(((.(((((.....))))).))).)).....-----.))))).......- ( -28.00) >DroEre_CAF1 8621 109 + 1 AGUGGGCACACACUAGUUGUGGCGACAUGGCAUUUUCUACGGAAGACGAAUACCGAAGGUUUAGAAAAUGCUUAGUUUGUAAAACCGAAAGUUCCGUUUUCAGAAUUUC- .((.(.((((.......)))).).))..((((((((((((....).((.....))......)))))))))))...((((.(((((.((....)).))))))))).....- ( -26.30) >DroYak_CAF1 11233 110 + 1 AGUGGGCACACACUAGUUGUGCCGAUAUGGCGUUUUCGCCGAAAAAGGAUUAUCAAAGAUUUAGAUAAUGCAAUGGUUAUAAAACCGAAAGCUCCGUUUUCAGAAAUACC .((.((((((.......)))))).))..((((....))))(((((.((((((((.........))))).((..(((((....)))))...))))).)))))......... ( -29.80) >consensus AGUGGGCACACACUAGUUGUGCCCACAUGGCAUUUUCGGCGAAAAACGAAUACCAAGGGUUUAGAAAAUGCAUAAUUUUUUAAACCAAAAG_____UUUUCAGAAUUUA_ .(((((((((.......)))))))))..............(((((............(((((((.((((.....)))).)))))))..........)))))......... (-13.89 = -16.17 + 2.28)

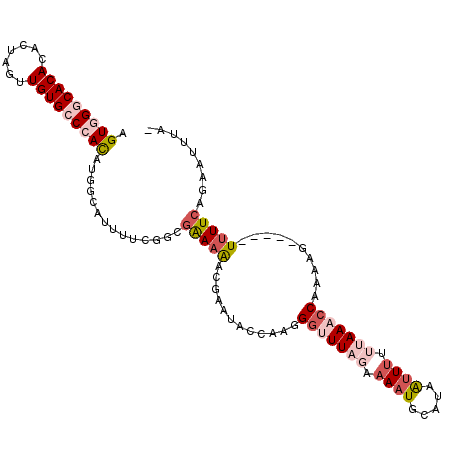
| Location | 21,755,979 – 21,756,083 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.80 |
| Mean single sequence MFE | -29.06 |
| Consensus MFE | -13.30 |
| Energy contribution | -15.42 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.18 |
| Structure conservation index | 0.46 |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.994453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21755979 104 - 22407834 -UAAAAACUGAAAA-----CUUUUGGUUUAAGAAAUGUUGCAUUUUCUAAACCCUUGGUAUUUGUUUUUUGACGAAAAUGCCAUGUGGCCACAACUAGUGUGUGCCCACU -.............-----.....((((((..(((((...)))))..))))))..(((((((((((....)))))..)))))).((((.((((.(....))))).)))). ( -26.40) >DroSec_CAF1 9562 104 - 1 -UAAAUUCUGAAAA-----CUUUUGGUUUAAAAAAUUAUGCAUUUUAUAAGCCCUUGGUUUUCGUUUUUCGCCGAAAAUUUCAUGUGGGCACAACUAGUGUGUGCCCACU -........(((((-----((...((((((.(((((.....))))).))))))...))))))).....................(((((((((.(....)))))))))). ( -31.00) >DroSim_CAF1 9571 103 - 1 -UAAAUUCUGAAAA-----CUUUUGGUUUAAAAAAUUAUGCAUUUUAUAAGCCCUUGGUUUUCGUU-UUCGCCGAAAAUUUCAUGUGGGCACAACUAGUGUGUGCCCACU -........(((((-----((...((((((.(((((.....))))).))))))...)))))))(((-(((...)))))).....(((((((((.(....)))))))))). ( -31.70) >DroEre_CAF1 8621 109 - 1 -GAAAUUCUGAAAACGGAACUUUCGGUUUUACAAACUAAGCAUUUUCUAAACCUUCGGUAUUCGUCUUCCGUAGAAAAUGCCAUGUCGCCACAACUAGUGUGUGCCCACU -(((((((((....))))).))))((.............((((((((((......((.....)).......))))))))))......((((((.....)))).))))... ( -24.12) >DroYak_CAF1 11233 110 - 1 GGUAUUUCUGAAAACGGAGCUUUCGGUUUUAUAACCAUUGCAUUAUCUAAAUCUUUGAUAAUCCUUUUUCGGCGAAAACGCCAUAUCGGCACAACUAGUGUGUGCCCACU (((((....(((((.(((((..(.((((....)))))..)).(((((.........)))))))).)))))((((....)))))))))((((((.(....))))))).... ( -32.10) >consensus _UAAAUUCUGAAAA_____CUUUUGGUUUAAAAAAUUAUGCAUUUUCUAAACCCUUGGUAUUCGUUUUUCGCCGAAAAUGCCAUGUGGGCACAACUAGUGUGUGCCCACU ........................((((((.(((((.....))))).))))))...............................(((((((((.(....)))))))))). (-13.30 = -15.42 + 2.12)

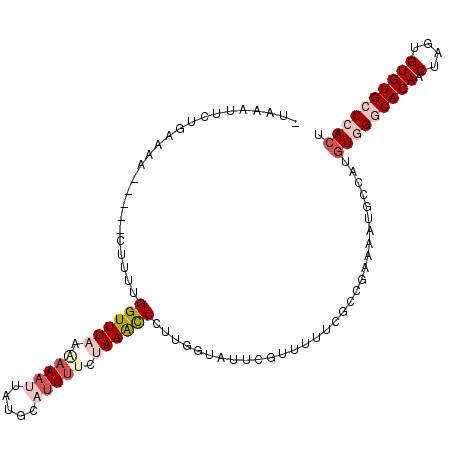
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:57 2006