
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,729,278 – 21,729,370 |
| Length | 92 |
| Max. P | 0.900038 |

| Location | 21,729,278 – 21,729,370 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 73.56 |
| Mean single sequence MFE | -21.08 |
| Consensus MFE | -13.69 |
| Energy contribution | -14.08 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.561361 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
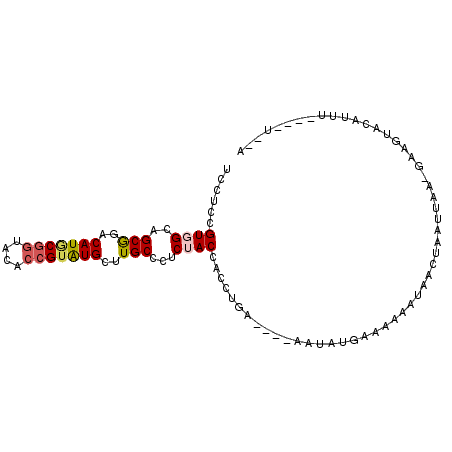
>2L_DroMel_CAF1 21729278 92 + 22407834 UCCUCCGUGGCAGCGGACAUGCGGUACACCGUAUGCUUGCCCUCUACCACCUGA----AAUAUGAAAAUAUAACUAAUUAA-GCAGUACAUUUUU--U--U ......((((.((.((.(((((((....))))))).....)).)).))))....----.((((....))))..........-.............--.--. ( -19.00) >DroVir_CAF1 5573 92 + 1 UCCUCCGUUGCGGCAGACAUUCGAUACACCGUGUGCUUGCCCUCCACCACCUGC----AGAUAGAGAAGCUAAUCAGUUUC-UGAGU----UGGGUAAAAU .......((((((((..(((.((......)).)))..)))).....(((.((.(----(((....((......))....))-)))).----)))))))... ( -20.80) >DroSim_CAF1 104 91 + 1 UCCUCCGUGGCAGCGGACAUGCGGUACACCGUAUGCUUGCCCUCUACCACCUGA----AAUAUGAAAAUAUAACUAAUUAA-GCAGUACAUUUU---U--U ......((((.((.((.(((((((....))))))).....)).)).))))....----.((((....))))..........-............---.--. ( -19.00) >DroEre_CAF1 120 91 + 1 UCCUCCGUGGCAGCGGACAUACGGUACACCGUAUGCUUGCCCUCUACCACCUGG----AAUAUAAGAAAAUAACUAACUGCACUGCUUCAGUU----U--A (((...((((.((.((.(((((((....))))))).....)).)).))))..))----)................(((((........)))))----.--. ( -23.00) >DroYak_CAF1 104 90 + 1 UCCUCCGUGGCAGCGGACAUACGGUACACCGUAUGCUUGCCCUCUACAACCUGG----AAUAGGAAAAAAUAACUAAUUAA-GAAGUACAUUC----U--A ...((((.((((..((.(((((((....))))))))))))))((((.....)))----)...)))...............(-(((.....)))----)--. ( -21.70) >DroAna_CAF1 120 100 + 1 UCCUCCGUGGCGGCGGACAUGCGGUACACAGUAUGCUUGCCCUCGACCACCUAAUGGUGAAAAGAAGAAAUAAUAUAUUAA-UAACAAGCUUCGAUUAAGA .....((.(((((((..(((((........)))))..))))......((((....))))......................-......))).))....... ( -23.00) >consensus UCCUCCGUGGCAGCGGACAUGCGGUACACCGUAUGCUUGCCCUCUACCACCUGA____AAUAUGAAAAAAUAACUAAUUAA_GAAGUACAUUU____U__A ......((((..(((..(((((((....)))))))..)))...))))...................................................... (-13.69 = -14.08 + 0.39)

| Location | 21,729,278 – 21,729,370 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 73.56 |
| Mean single sequence MFE | -26.15 |
| Consensus MFE | -17.74 |
| Energy contribution | -17.88 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.900038 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21729278 92 - 22407834 A--A--AAAAAUGUACUGC-UUAAUUAGUUAUAUUUUCAUAUU----UCAGGUGGUAGAGGGCAAGCAUACGGUGUACCGCAUGUCCGCUGCCACGGAGGA .--.--.(((((((((((.-.....))).))))))))....((----((..(((((((.(((((.((....((....)))).))))).))))))).)))). ( -28.00) >DroVir_CAF1 5573 92 - 1 AUUUUACCCA----ACUCA-GAAACUGAUUAGCUUCUCUAUCU----GCAGGUGGUGGAGGGCAAGCACACGGUGUAUCGAAUGUCUGCCGCAACGGAGGA .......((.----..(((-(...))))...((..(((((((.----......)))))))((((..((..(((....)))..))..))))))...)).... ( -23.90) >DroSim_CAF1 104 91 - 1 A--A---AAAAUGUACUGC-UUAAUUAGUUAUAUUUUCAUAUU----UCAGGUGGUAGAGGGCAAGCAUACGGUGUACCGCAUGUCCGCUGCCACGGAGGA .--.---(((((((((((.-.....))).))))))))....((----((..(((((((.(((((.((....((....)))).))))).))))))).)))). ( -28.00) >DroEre_CAF1 120 91 - 1 U--A----AACUGAAGCAGUGCAGUUAGUUAUUUUCUUAUAUU----CCAGGUGGUAGAGGGCAAGCAUACGGUGUACCGUAUGUCCGCUGCCACGGAGGA .--.----(((((........)))))...............((----((..(((((((.((....((((((((....)))))))))).))))))))))).. ( -31.30) >DroYak_CAF1 104 90 - 1 U--A----GAAUGUACUUC-UUAAUUAGUUAUUUUUUCCUAUU----CCAGGUUGUAGAGGGCAAGCAUACGGUGUACCGUAUGUCCGCUGCCACGGAGGA .--(----(((((.(((..-......))))))))).(((...(----((..((.((((.((....((((((((....)))))))))).)))).)))))))) ( -23.50) >DroAna_CAF1 120 100 - 1 UCUUAAUCGAAGCUUGUUA-UUAAUAUAUUAUUUCUUCUUUUCACCAUUAGGUGGUCGAGGGCAAGCAUACUGUGUACCGCAUGUCCGCCGCCACGGAGGA ((((.......((((((((-(....)))......((((...(((((....)))))..))))))))))...(((((....((......))...))))))))) ( -22.20) >consensus A__A____AAAUGUACUGC_UUAAUUAGUUAUUUUUUCAUAUU____UCAGGUGGUAGAGGGCAAGCAUACGGUGUACCGCAUGUCCGCUGCCACGGAGGA ...................................................(((((((.(((((.((............)).))))).)))))))...... (-17.74 = -17.88 + 0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:48 2006