
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,671,727 – 21,671,856 |
| Length | 129 |
| Max. P | 0.878205 |

| Location | 21,671,727 – 21,671,832 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 83.86 |
| Mean single sequence MFE | -32.72 |
| Consensus MFE | -19.91 |
| Energy contribution | -21.41 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
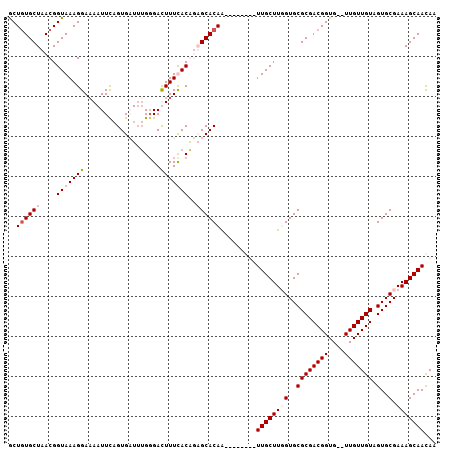
>2L_DroMel_CAF1 21671727 105 + 22407834 GCUGUGCUAACGGUAAAGGAAAAUUCAGUGAUUGGGGACUUUCACGUUGCACAGUUGAUGUAUUGCUUUGUGCGCGACGGUG--UUGUUGUAGUGCGAAAGCAACAA (((((((.(((.......((....)).((((..(....)..)))))))))))))).......((((((((..(((((((...--.)))))).)..).)))))))... ( -34.50) >DroSec_CAF1 5710 105 + 1 GCUGUGCUAACGGUAAAGGACAAUUUUGUGAUUUGGGACUUUCACGUUGCACAAUUAAUGUAUUGCUUUGUGCGCGACGGUG--UUGUUGUAGUGCGAAAGCAACAA ..(((((.((((..((((..(.(((....)))...)..))))..))))))))).........((((((((..(((((((...--.)))))).)..).)))))))... ( -29.60) >DroEre_CAF1 6033 99 + 1 ACUUUGCUAACGGUAAAGGAAAAUUCAGUGCUUUGGGAUUUUUACAGAGCACAA--------UUGCUUGGUGCGCGACGGUGCUUUGUUGUAGUGCGAAAGCAACAA .((((((.....))))))..........((((((.(.(((...(((((((((..--------((((.......))))..)))))))))...))).).)))))).... ( -34.10) >DroYak_CAF1 8307 98 + 1 GCUGUGCUAACGGUAAAGGAAAAUUCAGUGCUAUG-GAUUUUUACAGAGCACAA--------UUGCUUGGUGCGCGACGGUGCUUUGUUGUGGUGCGAAAGCAAUAA (((((....)))))....(((((((((......))-)))))))(((((((((..--------((((.......))))..))))))))).....(((....))).... ( -32.70) >consensus GCUGUGCUAACGGUAAAGGAAAAUUCAGUGAUUUGGGACUUUCACAGAGCACAA________UUGCUUGGUGCGCGACGGUG__UUGUUGUAGUGCGAAAGCAACAA ..((((((....(((((((...................)))))))..)))))).........((((((((..((((((((....))))))).)..).)))))))... (-19.91 = -21.41 + 1.50)


| Location | 21,671,763 – 21,671,856 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 75.13 |
| Mean single sequence MFE | -22.18 |
| Consensus MFE | -14.73 |
| Energy contribution | -15.04 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
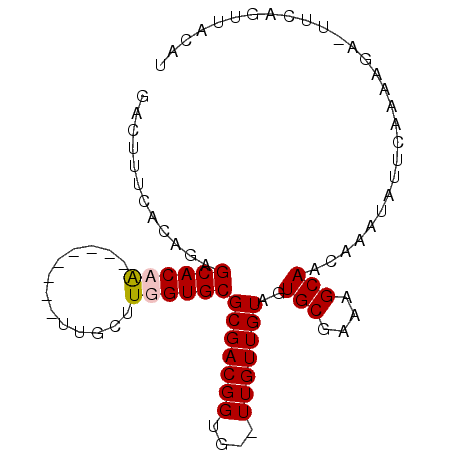
>2L_DroMel_CAF1 21671763 93 + 22407834 GACUUUCACGUUGCACAGUUGAUGUAUUGCUUUGUGCGCGACGGUG--UUGUUGUAGUGCGAAAGCAACAAAUAUUCAAAAAGAUUCAGUUAAAU ((((.(((((..((((.((((.(((((......))))))))).)))--)...)))..(((....)))...............))...)))).... ( -21.20) >DroSec_CAF1 5746 93 + 1 GACUUUCACGUUGCACAAUUAAUGUAUUGCUUUGUGCGCGACGGUG--UUGUUGUAGUGCGAAAGCAACAAAUAUUCAAAAGGUUUCAGUUACAU ((((((..(((((((((.....)))...((.....)))))))).((--.((((....(((....)))...))))..)).)))))).......... ( -20.00) >DroEre_CAF1 6069 86 + 1 GAUUUUUACAGAGCACAA--------UUGCUUGGUGCGCGACGGUGCUUUGUUGUAGUGCGAAAGCAACAAAUACGCAAAGGA-UAUAGUAACAU .(((((((((((((((..--------((((.......))))..))))))))).(((.(((....))).....)))...)))))-).......... ( -24.60) >DroYak_CAF1 8342 75 + 1 GAUUUUUACAGAGCACAA--------UUGCUUGGUGCGCGACGGUGCUUUGUUGUGGUGCGAAAGCAAUAAAUAUCCAAAGGA------------ (((.((((((((((((..--------((((.......))))..))))))))......(((....))).)))).))).......------------ ( -22.90) >consensus GACUUUCACAGAGCACAA________UUGCUUGGUGCGCGACGGUG__UUGUUGUAGUGCGAAAGCAACAAAUAUUCAAAAGA_UUCAGUUACAU ............((((((.............))))))(((((((....)))))))..(((....)))............................ (-14.73 = -15.04 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:45 2006