
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,623,200 – 21,623,290 |
| Length | 90 |
| Max. P | 0.889136 |

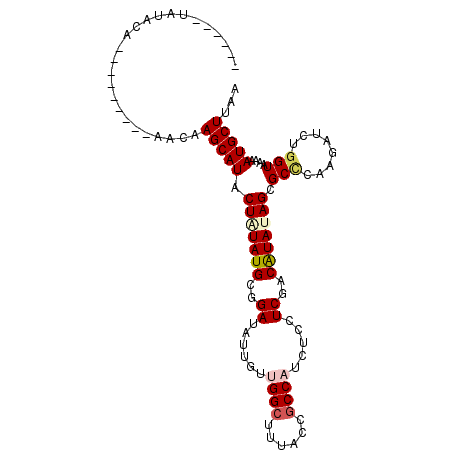
| Location | 21,623,200 – 21,623,290 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 78.72 |
| Mean single sequence MFE | -19.85 |
| Consensus MFE | -14.24 |
| Energy contribution | -14.80 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745711 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21623200 90 + 22407834 ------UGUUCA---------AACAAGCAUUCUAUAUGCGGAUAUCGUUGGCUUUACCGCCAUCUCCUCCACAUAUAGUGCGCAAGAUUUGGUAAAAAUGCUUAA ------......---------...((((((((((((((.(((......((((......)))).....))).)))))))..(....)..........))))))).. ( -24.00) >DroVir_CAF1 33012 96 + 1 AAAAUAUAUAUA---------UAUAAGCAUACUGUAUGCGGAUAUUGUUGGGUUUACCGCCAUCUCCUCGACGUAUAGCGCCCAGGAUCUGGUAAAAAUGCUAAA ............---------....(((((.(((....)))........((.....))((((..((((.(.((.....)).).))))..))))....)))))... ( -19.70) >DroWil_CAF1 29606 99 + 1 ------UACACACACACCAUCAAAUAGCAUACUUUAUGCGGAUAUUGUUGGCUUUACUGCCAUCUCCUCAACAUAUAGCGCCCAAGAUCUGGUUAAAAUGCUUAA ------...................(((((.((.((((..((....(.((((......)))).)...))..)))).))...(((.....))).....)))))... ( -16.40) >DroMoj_CAF1 22238 82 + 1 AA-----------------------AGCAUACUGUAUGCGGAUAUUGUCGGCUUUACCGCCAUCUCCUCGACGUAUAGCGCCCAAGAUCUGGUAAAAAUGCUAAA ..-----------------------(((((.((((((((((.....(..(((......)))..)...))).))))))).(((........)))....)))))... ( -19.00) >DroAna_CAF1 16694 90 + 1 ------CGUGAA---------AACAAGCAUACUCUAUGCGGAUAUCGUUGGCUUCACAGCCAUCUCCUCGACAUAUAGCGCCCAAGAUUUGGUUAAAAUGCUUAA ------.((...---------.))((((((.((.(((((((.......(((((....))))).....))).)))).))...(((.....))).....)))))).. ( -18.80) >DroPer_CAF1 33183 90 + 1 ------UUUACG---------AAUAAGCAUACUAUAUGCGGAUAUAGUGGGCUUUACCGCCAUCUCCUCGACAUAUAGCGCUCAAGACUUGGUUAAAAUGCUUAA ------......---------..(((((((.(((((((((((.((.((((......)))).)).)))..).))))))).(((........)))....))))))). ( -21.20) >consensus ______UAUACA_________AACAAGCAUACUAUAUGCGGAUAUUGUUGGCUUUACCGCCAUCUCCUCGACAUAUAGCGCCCAAGAUCUGGUAAAAAUGCUUAA .........................(((((.(((((((..((......((((......)))).....))..))))))).(((........)))....)))))... (-14.24 = -14.80 + 0.56)


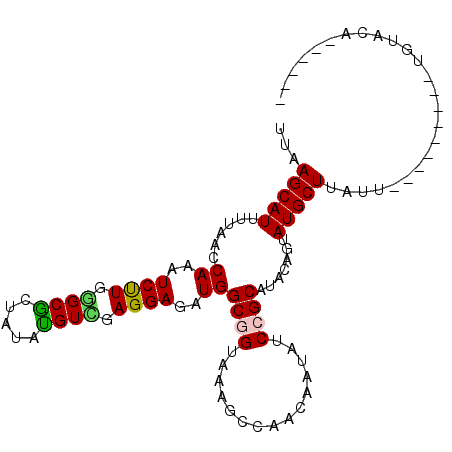
| Location | 21,623,200 – 21,623,290 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 78.72 |
| Mean single sequence MFE | -24.35 |
| Consensus MFE | -16.58 |
| Energy contribution | -16.28 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.889136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21623200 90 - 22407834 UUAAGCAUUUUUACCAAAUCUUGCGCACUAUAUGUGGAGGAGAUGGCGGUAAAGCCAACGAUAUCCGCAUAUAGAAUGCUUGUU---------UGAACA------ ..............(((((...(((..(((((((((((...(.((((......)))).)....)))))))))))..)))..)))---------))....------ ( -29.20) >DroVir_CAF1 33012 96 - 1 UUUAGCAUUUUUACCAGAUCCUGGGCGCUAUACGUCGAGGAGAUGGCGGUAAACCCAACAAUAUCCGCAUACAGUAUGCUUAUA---------UAUAUAUAUUUU ...(((((......((..((((.((((.....)))).))))..))((((...............)))).......)))))....---------............ ( -23.16) >DroWil_CAF1 29606 99 - 1 UUAAGCAUUUUAACCAGAUCUUGGGCGCUAUAUGUUGAGGAGAUGGCAGUAAAGCCAACAAUAUCCGCAUAAAGUAUGCUAUUUGAUGGUGUGUGUGUA------ ....((((....((((.......((((((.(((((.((...(.((((......)))).)....)).))))).)))..)))......))))....)))).------ ( -24.62) >DroMoj_CAF1 22238 82 - 1 UUUAGCAUUUUUACCAGAUCUUGGGCGCUAUACGUCGAGGAGAUGGCGGUAAAGCCGACAAUAUCCGCAUACAGUAUGCU-----------------------UU ...(((((.....(((..((((.((((.....)))).))))..)))(((.....)))..................)))))-----------------------.. ( -21.30) >DroAna_CAF1 16694 90 - 1 UUAAGCAUUUUAACCAAAUCUUGGGCGCUAUAUGUCGAGGAGAUGGCUGUGAAGCCAACGAUAUCCGCAUAGAGUAUGCUUGUU---------UUCACG------ .(((((((.....(((.....)))..(((.(((((.((...(.(((((....))))).)....)).))))).))))))))))..---------......------ ( -24.10) >DroPer_CAF1 33183 90 - 1 UUAAGCAUUUUAACCAAGUCUUGAGCGCUAUAUGUCGAGGAGAUGGCGGUAAAGCCCACUAUAUCCGCAUAUAGUAUGCUUAUU---------CGUAAA------ .(((((((.........((.....))(((((((((...(((.((((.((......)).)))).)))))))))))))))))))..---------......------ ( -23.70) >consensus UUAAGCAUUUUAACCAAAUCUUGGGCGCUAUAUGUCGAGGAGAUGGCGGUAAAGCCAACAAUAUCCGCAUACAGUAUGCUUAUU_________UGUACA______ ...(((((......((..((((.((((.....)))).))))..))((((...............)))).......)))))......................... (-16.58 = -16.28 + -0.30)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:35 2006