
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,616,302 – 21,616,402 |
| Length | 100 |
| Max. P | 0.977385 |

| Location | 21,616,302 – 21,616,402 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 72.08 |
| Mean single sequence MFE | -23.49 |
| Consensus MFE | -10.15 |
| Energy contribution | -9.35 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.43 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
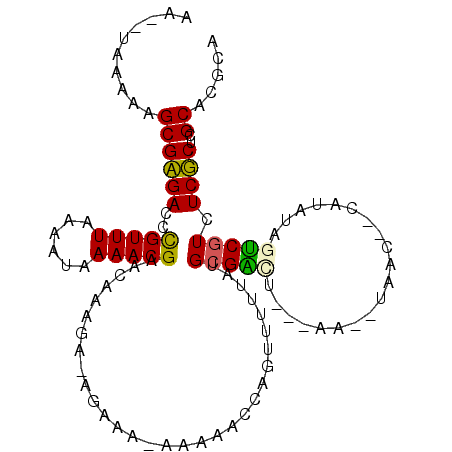
>2L_DroMel_CAF1 21616302 100 + 22407834 AA--UGAAAAGCGAGACCCGUUUAAACUAAAACGAACAAAGAUAGAAAUAAAAACCAGUUUUAAUGGGAUU---AA--UGAC--AAUAUAGUCGUUUCUCUCGCACUCA ..--......((((((((((((.(((((....(.......).((....))......))))).))))))...---((--((((--......))))))..))))))..... ( -23.70) >DroPse_CAF1 10445 99 + 1 AG--UAAAAAGCGCGACUUGUUUGAAAUAAAACGAACGAA------AAAAAAAACUGGUUUUUAUGCGGCGCGCACACAAGU--CAUACAUCCGUCUCGCUUGCGCGCA ..--.......((((..(((((((........))))))).------...(((((....))))).))))(((((((..(.((.--(........).)).)..))))))). ( -23.90) >DroEre_CAF1 10617 97 + 1 AA--UGAAAAGCGAGACCCGUUUAAAUUAAAACGAACAAAGAGAGAAA---AAACCAGUUUUCAGGCGACU---AA--UGAC--AAUAUAGUCGUUUCUCUCGCACGCA ..--......(((.....(((((......)))))......((((((((---...((........))(((((---((--(...--.)).))))))))))))))...))). ( -23.90) >DroYak_CAF1 10568 98 + 1 AA--UGAAAAGCGAGACCCGUUUAAACUAAAACGAACAAAGAGAGAAA--AAAACCAGUUUUUAUGCGACU---AA--UGUC--CAUAUAGUCGUUUCUCUCGCACGCA ..--......(((.....(((((......)))))......((((((((--((((....)))))..((((((---(.--....--....))))))))))))))...))). ( -24.30) >DroAna_CAF1 8940 105 + 1 AAAGUAAAAAGCGCGACUUGUUUGAAAUUAAACGAACCCGAACGAAGAGAAAAACUGGUUUUUAUGCGGCU---ACA-UAACCCCCAAUAGUCGUCUCGCUUGCACACG ...((...(((((.((((((((((..............)))))))...((.....(((...(((((.....---.))-)))...)))....))))).)))))..))... ( -20.24) >DroPer_CAF1 25427 98 + 1 AG--UAAAAAGCGCGACUUGUUUGAAAUAAAACGAACGAA------AA-AAAAACUGGUUUUUAUGCGGCGCGCACACAAGC--CAUACAUCCGUCUCGCUUGCGCGCA .(--(...(((((.((((((((((........))))))).------..-......((((((.(.((((...)))).).))))--)).......))).)))))..))... ( -24.90) >consensus AA__UAAAAAGCGAGACCCGUUUAAAAUAAAACGAACAAAGA_AGAAA_AAAAACCAGUUUUUAUGCGACU___AA__UAAC__CAUAUAGUCGUCUCGCUCGCACGCA ..........((((((..(((((......)))))...............................(((((....................))))).))))..))..... (-10.15 = -9.35 + -0.80)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:32 2006