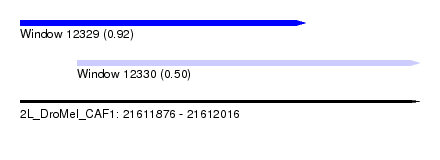
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,611,876 – 21,612,016 |
| Length | 140 |
| Max. P | 0.917411 |

| Location | 21,611,876 – 21,611,976 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 75.71 |
| Mean single sequence MFE | -20.23 |
| Consensus MFE | -12.62 |
| Energy contribution | -14.73 |
| Covariance contribution | 2.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.917411 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
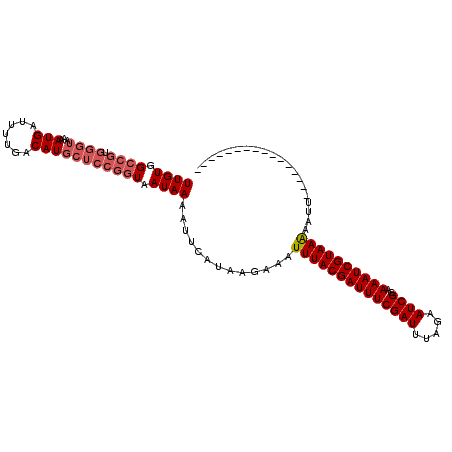
>2L_DroMel_CAF1 21611876 100 + 22407834 UUGUGGCGC--------AUGA---UAACAUGAUCCGAUAAUAAAUUUCGUAAAAACUUUACGAUUUCGAUUUAAAAUCGCAAAAUCGUAAGAAUUUUUUUAUUUAUAAAAC ...(((..(--------(((.---...))))..)))...((((((.....((((..((((((((((((((.....))))..))))))))))..))))...))))))..... ( -17.70) >DroSec_CAF1 71064 95 + 1 UUGUGGCCGUGGGUAAAAUGAUUUUGACAUGCUCCGGUAAUAAAAUUCAUAAGAAUUUUACGAUUUCGAUUUAGAAUCGCAAAAUCGUAAAAAUU---------------- ((((.((((.(((((..............))))))))).))))............(((((((((((((((.....))))..)))))))))))...---------------- ( -21.94) >DroSim_CAF1 10111 95 + 1 UUGUGGCCGUGGGUAAAAUGAUUUUGACAUGCUCCGGUAAUAAAAUUCAUAAGAAAUUUACGAUUUCGAUUUAGAAUCGCAAAAUCGUAAAAAUU---------------- ((((.((((.(((((..............))))))))).)))).............((((((((((((((.....))))..))))))))))....---------------- ( -21.04) >consensus UUGUGGCCGUGGGUAAAAUGAUUUUGACAUGCUCCGGUAAUAAAAUUCAUAAGAAAUUUACGAUUUCGAUUUAGAAUCGCAAAAUCGUAAAAAUU________________ ((((.((((.((((...(((.......))))))))))).)))).............((((((((((((((.....))))..)))))))))).................... (-12.62 = -14.73 + 2.11)

| Location | 21,611,896 – 21,612,016 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.35 |
| Mean single sequence MFE | -21.21 |
| Consensus MFE | -16.69 |
| Energy contribution | -16.03 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
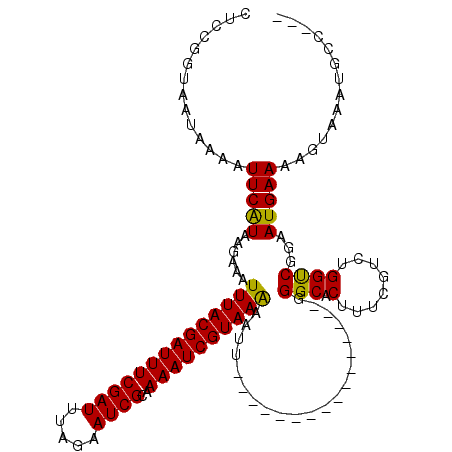
>2L_DroMel_CAF1 21611896 120 + 22407834 AUCCGAUAAUAAAUUUCGUAAAAACUUUACGAUUUCGAUUUAAAAUCGCAAAAUCGUAAGAAUUUUUUUAUUUAUAAAACGGGCACUUUUGAAUGGCCGUAAUGAAAAGUAAAUGCCUGU ....(((((.(((.(((((....)).(((((((((((((.....))))..)))))))))))).))).)))))......((((((((((((.(.((....)).).))))))....)))))) ( -22.20) >DroSec_CAF1 71095 101 + 1 CUCCGGUAAUAAAAUUCAUAAGAAUUUUACGAUUUCGAUUUAGAAUCGCAAAAUCGUAAAAAUU----------------UGGCACUUUCGUCUGGUCGGAAUGAAAAGUAAAUGCC--- ....((((......(((((.....(((((((((((((((.....))))..))))))))))).((----------------((((.(........)))))))))))).......))))--- ( -20.32) >DroSim_CAF1 10142 101 + 1 CUCCGGUAAUAAAAUUCAUAAGAAAUUUACGAUUUCGAUUUAGAAUCGCAAAAUCGUAAAAAUU----------------GGGCACUUUCGUCUGGUCGGAAUGAAAAGUAAAUGCC--- .((((....................((((((((((((((.....))))..)))))))))).((.----------------.(((......)))..))))))................--- ( -21.10) >consensus CUCCGGUAAUAAAAUUCAUAAGAAAUUUACGAUUUCGAUUUAGAAUCGCAAAAUCGUAAAAAUU________________GGGCACUUUCGUCUGGUCGGAAUGAAAAGUAAAUGCC___ ..............(((((......((((((((((((((.....))))..)))))))))).....................(((.(........))))...))))).............. (-16.69 = -16.03 + -0.66)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:30 2006