| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,561,078 – 21,561,183 |
| Length | 105 |
| Max. P | 0.507679 |

| Location | 21,561,078 – 21,561,183 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.02 |
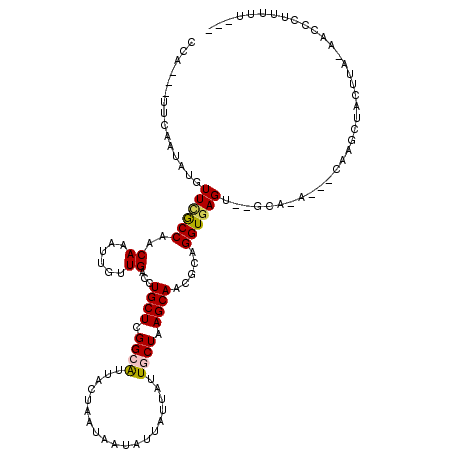
| Mean single sequence MFE | -24.75 |
| Consensus MFE | -9.65 |
| Energy contribution | -9.63 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.39 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
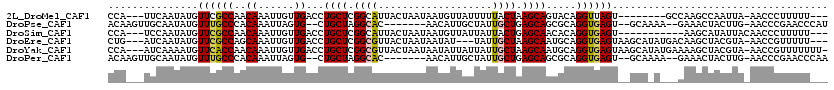
>2L_DroMel_CAF1 21561078 105 + 22407834 CCA---UUCAAUAUGUUCGCCAACAAAUUGUUGACCUGCUCGGCAUUACUAAUAAUGUUAUUUUUACUAAGCAGUACAGGUUAGU--------GCCAAGCCAAUUA-AACCCUUUUU--- .((---..((((.((((....)))).)))).))....(((.(((((((((.....((((..........)))).....)).))))--------))).)))......-..........--- ( -17.80) >DroPse_CAF1 82155 106 + 1 ACAAGUUGCAAUAUGUUUGCCCACAAAUUAGUG--CUGCUAGGCAC-------AACAUUGCUAUUGCUGAGCAGCGCAGGUGAGU--GCAAAA--GAAACUACUUG-AACCCGAACCCAU .(((((.........(((((((((......(((--(((((.((((.-------...........)))).))))))))..))).).--))))).--......)))))-............. ( -29.09) >DroSim_CAF1 141530 103 + 1 CCA---UCCAAUAUGUUCGCCAACAAAUUGUUGACCUGCUCGGCAUUACUAAUAAUGUUAUUAUUACUGAGCAACACAGGUGAGU-----------AAGCAUAUUACAACCCUUUUU--- ...---...((((((((..(((((.....))))(((((....((.(((.(((((((...))))))).)))))....)))))..).-----------.))))))))............--- ( -22.60) >DroEre_CAF1 197049 110 + 1 CUG---AUCAAUAUGUUCGCCAGCAAAUUGUUGACCUGCUCGGCGUUACUAAUAAUAU---UAUUGCUAAGCAAUGCAGGUGAGUAAGCAUAUGACAAGCUACGUA-AACCGUUUUU--- ((.---.(((.((((((..(((((.....))))((((((...((.(((.(((((....---))))).)))))...))))))..)..)))))))))..)).......-..........--- ( -25.80) >DroYak_CAF1 170077 115 + 1 CCA---AUCAAAAUGUUCACCAACAAAUUGUUGACCUGCUCGGCGUUACUAAUAAUAUUAUUAUUGCUAAGCAAUGCAGGUGAGUAAGCAUAUGAAAAGCUACGUA-AACCGUUUUUUU- ...---......(((((.((((((.....))))((((((...((.(((.(((((((...))))))).)))))...))))))..)).)))))..(((((((......-....))))))).- ( -24.10) >DroPer_CAF1 80812 106 + 1 ACAAGUUGCAAUAUGUUUGCCCACAAAUUAGUG--CUGCUAGGCAC-------AACAUUGCUAUUGCUGAGCAGCGCAGGUGAGU--GCAAAA--GAAACUACUUG-AACCCGAACCCAA .(((((.........(((((((((......(((--(((((.((((.-------...........)))).))))))))..))).).--))))).--......)))))-............. ( -29.09) >consensus CCA___UUCAAUAUGUUCGCCAACAAAUUGUUGACCUGCUCGGCAUUACUAAUAAUAUUAUUAUUGCUAAGCAACGCAGGUGAGU__GCA_A___CAAGCUACUUA_AACCCUUUUU___ ...............((((((..((......))...((((.((((...................)))).)))).....)))))).................................... ( -9.65 = -9.63 + -0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:27 2006