
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,490,423 – 21,490,579 |
| Length | 156 |
| Max. P | 0.999961 |

| Location | 21,490,423 – 21,490,542 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.47 |
| Mean single sequence MFE | -31.30 |
| Consensus MFE | -14.57 |
| Energy contribution | -16.07 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.47 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962677 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21490423 119 + 22407834 AAUUACCCAUAUUGCGGGUAAAUAAGUUUUAUUUGUAAAUUCAUAUUCGAUGAUUGGUG-GUUGAAAAAUGCAUUUCUUUGGUAUAACACAUUGUGGCCCUGAAAAGGGCCGUUUUGGAU ..((((((.......))))))....(((.((((.(.((((.(((.((((((.(....).-))))))..))).)))))...)))).))).((..((((((((....))))))))..))... ( -30.30) >DroEre_CAF1 171928 116 + 1 -UUUACCCAUAUUAUCGGUAG---AGUUAAAAUGGUAAAUUCAUAUUAGACGAUUGGUAAAUUGAAAAAUACAUUUCUUUGAUGAAACACAUUGUGGCCCUGAAAAGGGCCGUUUUGGAU -.....(((..(((((((.((---((((((.((((.....)))).)))).(((((....)))))..........)))))))))))..........((((((....))))))....))).. ( -27.40) >DroYak_CAF1 159390 104 + 1 ---------UUUUAACACCAU---AGUGGGAAUGGAUAAGUUGAAAUCGACGA----UAUAUUGAAAAAUACAUUUCCUUGGUGAAACACAUUGUGGCCCUGAAAAGGGCCGUUUUGGAU ---------......(((((.---...(((((((.....((((....))))..----..((((....))))))))))).))))).....((..((((((((....))))))))..))... ( -36.20) >consensus __UUACCCAUAUUAACGGUAA___AGUUAAAAUGGUAAAUUCAUAUUCGACGAUUGGUA_AUUGAAAAAUACAUUUCUUUGGUGAAACACAUUGUGGCCCUGAAAAGGGCCGUUUUGGAU ...............................................................((((......))))............((..((((((((....))))))))..))... (-14.57 = -16.07 + 1.50)


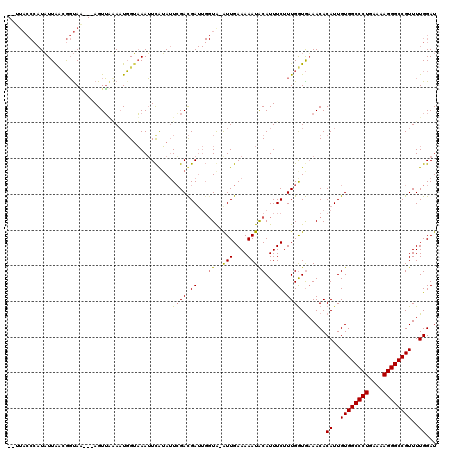
| Location | 21,490,423 – 21,490,542 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.47 |
| Mean single sequence MFE | -22.70 |
| Consensus MFE | -16.67 |
| Energy contribution | -16.57 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.73 |
| SVM decision value | 4.91 |
| SVM RNA-class probability | 0.999961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21490423 119 - 22407834 AUCCAAAACGGCCCUUUUCAGGGCCACAAUGUGUUAUACCAAAGAAAUGCAUUUUUCAAC-CACCAAUCAUCGAAUAUGAAUUUACAAAUAAAACUUAUUUACCCGCAAUAUGGGUAAUU .........((((((....))))))....((((..........((((......))))...-......((((.....))))...))))............(((((((.....))))))).. ( -23.10) >DroEre_CAF1 171928 116 - 1 AUCCAAAACGGCCCUUUUCAGGGCCACAAUGUGUUUCAUCAAAGAAAUGUAUUUUUCAAUUUACCAAUCGUCUAAUAUGAAUUUACCAUUUUAACU---CUACCGAUAAUAUGGGUAAA- .........((((((....))))))..((((..((((......))))..))))......((((((..((((.....))))......(((.(((.(.---.....).))).)))))))))- ( -23.30) >DroYak_CAF1 159390 104 - 1 AUCCAAAACGGCCCUUUUCAGGGCCACAAUGUGUUUCACCAAGGAAAUGUAUUUUUCAAUAUA----UCGUCGAUUUCAACUUAUCCAUUCCCACU---AUGGUGUUAAAA--------- .........((((((....))))))...........(((((.((..(((((((....))))))----)..))(((........)))..........---.)))))......--------- ( -21.70) >consensus AUCCAAAACGGCCCUUUUCAGGGCCACAAUGUGUUUCACCAAAGAAAUGUAUUUUUCAAU_UACCAAUCGUCGAAUAUGAAUUUACCAUUAAAACU___AUACCGAUAAUAUGGGUAA__ .........((((((....))))))..((((..((((......))))..))))................................................................... (-16.67 = -16.57 + -0.11)

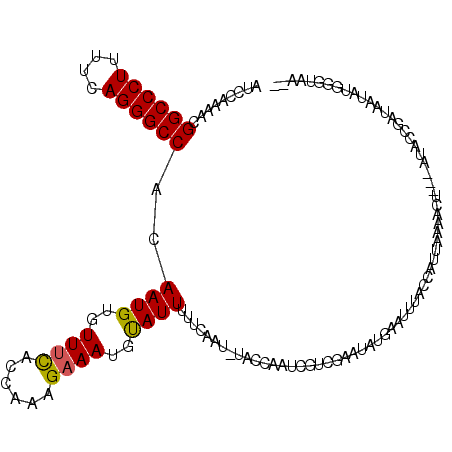

| Location | 21,490,463 – 21,490,579 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.63 |
| Mean single sequence MFE | -31.33 |
| Consensus MFE | -20.39 |
| Energy contribution | -21.12 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.625618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21490463 116 + 22407834 UCAUAUUCGAUGAUUGGUG-GUUGAAAAAUGCAUUUCUUUGGUAUAACACAUUGUGGCCCUGAAAAGGGCCGUUUUGGAUUAUUGUCCGC---AUUCGCAGGAGAAAAUUAUUUAGAGCU .......(((((....(((-.(.((((......))))...).)))....))))).((((((....))))))(((((((((..((.(((((---....)).))).))....))))))))). ( -30.60) >DroEre_CAF1 171964 117 + 1 UCAUAUUAGACGAUUGGUAAAUUGAAAAAUACAUUUCUUUGAUGAAACACAUUGUGGCCCUGAAAAGGGCCGUUUUGGAUUUUUGUCCGC---ACUCGCAGGAGGAAAUUAUUUGGAGCU ...(((((((.((...(((..((....)))))...)))))))))...........((((((....))))))((((..(((((((.(((((---....)).))).))))..)))..)))). ( -33.50) >DroYak_CAF1 159418 116 + 1 UUGAAAUCGACGA----UAUAUUGAAAAAUACAUUUCCUUGGUGAAACACAUUGUGGCCCUGAAAAGGGCCGUUUUGGAUUAAUGUCCGCAGCAUUAGUAGAAACAAAUUAUUUGGAGCU ...........((----(.((((....)))).)))(((..(((((..........((((((....))))))(((((..((((((((.....)))))))).)))))...))))).)))... ( -29.90) >consensus UCAUAUUCGACGAUUGGUA_AUUGAAAAAUACAUUUCUUUGGUGAAACACAUUGUGGCCCUGAAAAGGGCCGUUUUGGAUUAUUGUCCGC___AUUCGCAGGAGAAAAUUAUUUGGAGCU ..................................((((..(((((......((((((((((....)))))).....((((....)))).........)))).......))))).)))).. (-20.39 = -21.12 + 0.73)

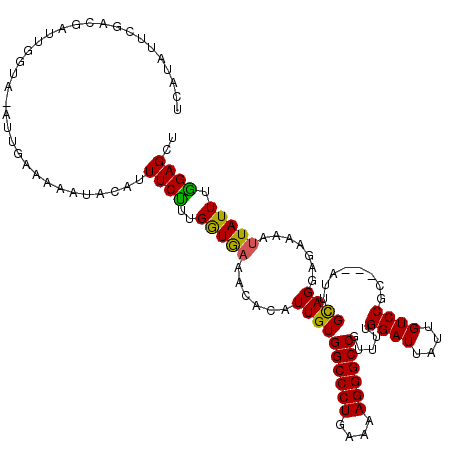
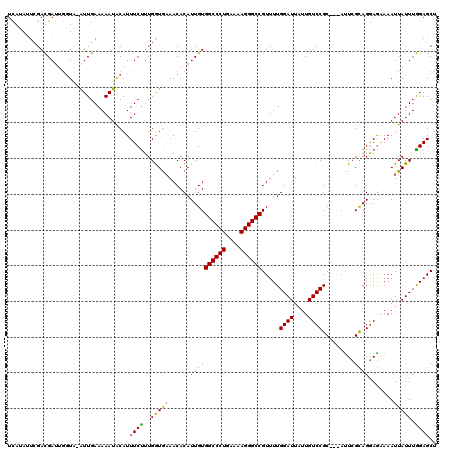
| Location | 21,490,463 – 21,490,579 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.63 |
| Mean single sequence MFE | -21.98 |
| Consensus MFE | -19.07 |
| Energy contribution | -18.97 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620523 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21490463 116 - 22407834 AGCUCUAAAUAAUUUUCUCCUGCGAAU---GCGGACAAUAAUCCAAAACGGCCCUUUUCAGGGCCACAAUGUGUUAUACCAAAGAAAUGCAUUUUUCAAC-CACCAAUCAUCGAAUAUGA .((..................))((((---(((((......))).....((((((....)))))).......................))))))......-................... ( -20.27) >DroEre_CAF1 171964 117 - 1 AGCUCCAAAUAAUUUCCUCCUGCGAGU---GCGGACAAAAAUCCAAAACGGCCCUUUUCAGGGCCACAAUGUGUUUCAUCAAAGAAAUGUAUUUUUCAAUUUACCAAUCGUCUAAUAUGA .((((((.............)).))))---..((((.............((((((....))))))..((((..((((......))))..))))................))))....... ( -24.62) >DroYak_CAF1 159418 116 - 1 AGCUCCAAAUAAUUUGUUUCUACUAAUGCUGCGGACAUUAAUCCAAAACGGCCCUUUUCAGGGCCACAAUGUGUUUCACCAAGGAAAUGUAUUUUUCAAUAUA----UCGUCGAUUUCAA (((........................)))...(((.............((((((....))))))..((((..((((......))))..))))..........----..)))........ ( -21.06) >consensus AGCUCCAAAUAAUUUCCUCCUGCGAAU___GCGGACAAUAAUCCAAAACGGCCCUUUUCAGGGCCACAAUGUGUUUCACCAAAGAAAUGUAUUUUUCAAU_UACCAAUCGUCGAAUAUGA ................................(((......))).....((((((....))))))..((((..((((......))))..))))........................... (-19.07 = -18.97 + -0.11)


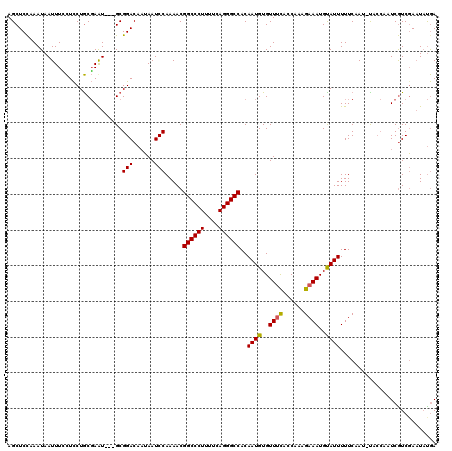
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:23 2006