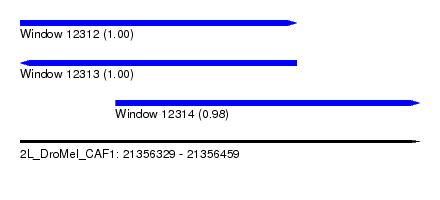
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,356,329 – 21,356,459 |
| Length | 130 |
| Max. P | 0.999405 |

| Location | 21,356,329 – 21,356,419 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 77.86 |
| Mean single sequence MFE | -20.05 |
| Consensus MFE | -14.70 |
| Energy contribution | -15.64 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.73 |
| SVM decision value | 3.32 |
| SVM RNA-class probability | 0.998990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
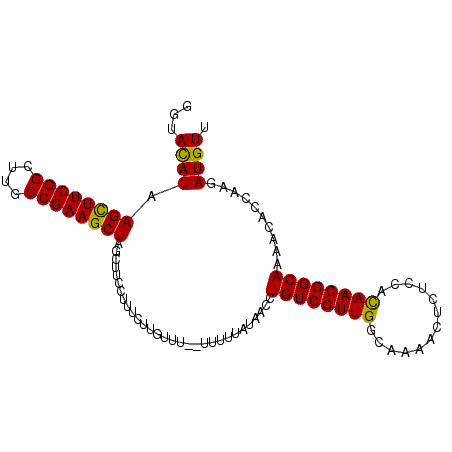
>2L_DroMel_CAF1 21356329 90 + 22407834 UC-U-U--UUUUUUCAUAAAUUAAAUUGUUGCAAGGGUAUAUAAGCUUCGGCUUGCCGAAGCUAGCUUCCUUUCUUGUUU---UUUUAUAACCUGUC ..-.-.--.......(((((..((((....(.(((((......((((((((....))))))))....))))).)..))))---.)))))........ ( -18.80) >DroSec_CAF1 65761 79 + 1 ----------------UAAAUUAAAUUUUUGCAAGGGUACAUAAGCUUCGGCUUGCCGAAGCUAGCUUCCUUCCUUGUUU--UUUUUAUAACCUGUC ----------------.....((((.....(((((((......((((((((....))))))))........)))))))..--..))))......... ( -21.74) >DroEre_CAF1 48794 84 + 1 UUGG-UA--UUUUUCUUAAAUAAAAUCAU-----AAGUACAUAAGUUUCGGCUUGCCGAAGCUAGCUU----UGUUGAUUU-UUUUUAUAACCUGUC ..((-(.--.......((((.(((((((.-----((((.....((((((((....)))))))).))))----...))))))-).))))..))).... ( -18.80) >DroYak_CAF1 52823 97 + 1 UUGGGUAUUUUUUUCUUAAAUUAAAUUAUUGCAAGGGUACAUAAGCUUCGGCUUGCCGAAGCUAGCUUCCUUUCUUGUUUUGUUUUUAUAACCUGUC ..((((((((.......))))((((.((..(((((((......((((((((....))))))))........)))))))..))..))))..))))... ( -20.84) >consensus UU_G_U___UUUUUCUUAAAUUAAAUUAUUGCAAGGGUACAUAAGCUUCGGCUUGCCGAAGCUAGCUUCCUUUCUUGUUU__UUUUUAUAACCUGUC ..............................(((((((......((((((((....))))))))........)))))))................... (-14.70 = -15.64 + 0.94)

| Location | 21,356,329 – 21,356,419 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 77.86 |
| Mean single sequence MFE | -18.40 |
| Consensus MFE | -13.40 |
| Energy contribution | -14.78 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.73 |
| SVM decision value | 3.57 |
| SVM RNA-class probability | 0.999405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21356329 90 - 22407834 GACAGGUUAUAAAA---AAACAAGAAAGGAAGCUAGCUUCGGCAAGCCGAAGCUUAUAUACCCUUGCAACAAUUUAAUUUAUGAAAAAA--A-A-GA ......(((((((.---........((((.....((((((((....)))))))).......))))............))))))).....--.-.-.. ( -17.90) >DroSec_CAF1 65761 79 - 1 GACAGGUUAUAAAAA--AAACAAGGAAGGAAGCUAGCUUCGGCAAGCCGAAGCUUAUGUACCCUUGCAAAAAUUUAAUUUA---------------- ...............--......(.((((..((.((((((((....))))))))...))..)))).)..............---------------- ( -21.30) >DroEre_CAF1 48794 84 - 1 GACAGGUUAUAAAAA-AAAUCAACA----AAGCUAGCUUCGGCAAGCCGAAACUUAUGUACUU-----AUGAUUUUAUUUAAGAAAAA--UA-CCAA ....(((..((((.(-((((((...----..((.((.(((((....))))).))...))....-----.))))))).)))).......--.)-)).. ( -14.30) >DroYak_CAF1 52823 97 - 1 GACAGGUUAUAAAAACAAAACAAGAAAGGAAGCUAGCUUCGGCAAGCCGAAGCUUAUGUACCCUUGCAAUAAUUUAAUUUAAGAAAAAAAUACCCAA ....((.(((.............(.((((..((.((((((((....))))))))...))..)))).)..(((......)))........))).)).. ( -20.10) >consensus GACAGGUUAUAAAAA__AAACAAGAAAGGAAGCUAGCUUCGGCAAGCCGAAGCUUAUGUACCCUUGCAAUAAUUUAAUUUAAGAAAAA___A_C_AA .......................(.((((..((.((((((((....))))))))...))..)))).).............................. (-13.40 = -14.78 + 1.38)


| Location | 21,356,360 – 21,356,459 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 92.27 |
| Mean single sequence MFE | -24.84 |
| Consensus MFE | -23.66 |
| Energy contribution | -23.09 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975282 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21356360 99 + 22407834 GGUAUAUAAGCUUCGGCUUGCCGAAGCUAGCUUCCUUUCUUGUUU---UUUUAUAACCUGUCGUUGGCAAAACUCUCCACAACGGCAAAACACUAAGAUGUU (((.((((((((((((....))))))))(((..........))).---...)))))))((((((((.............))))))))............... ( -25.42) >DroSec_CAF1 65780 100 + 1 GGUACAUAAGCUUCGGCUUGCCGAAGCUAGCUUCCUUCCUUGUUU--UUUUUAUAACCUGUCGUUGGCAAAACUCUCCACAACGGCAAAACACUAAGAUGUU ...((((.((((((((....))))))))......(((...(((((--...........((((((((.............)))))))))))))..))))))). ( -25.22) >DroEre_CAF1 48821 97 + 1 AGUACAUAAGUUUCGGCUUGCCGAAGCUAGCUU----UGUUGAUUU-UUUUUAUAACCUGUCGUUGGCAAAACUCUCCACAACGGCAAAACACCAAGAUGUU ...((((.((((((((....)))))))).....----.........-...........((((((((.............))))))))..........)))). ( -22.42) >DroYak_CAF1 52858 102 + 1 GGUACAUAAGCUUCGGCUUGCCGAAGCUAGCUUCCUUUCUUGUUUUGUUUUUAUAACCUGUCGUUGGCAAAACUCUCCAUAACGGCAAAACACCAAGAUGUU ((......((((((((....)))))))).....))..(((((...(((((........((((((((.............))))))))))))).))))).... ( -26.32) >consensus GGUACAUAAGCUUCGGCUUGCCGAAGCUAGCUUCCUUUCUUGUUU__UUUUUAUAACCUGUCGUUGGCAAAACUCUCCACAACGGCAAAACACCAAGAUGUU ...((((.((((((((....))))))))..............................((((((((.............))))))))..........)))). (-23.66 = -23.09 + -0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:16 2006