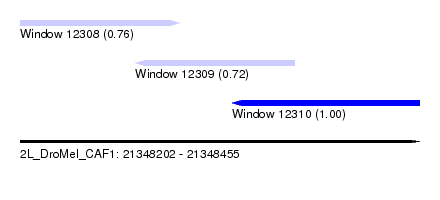
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,348,202 – 21,348,455 |
| Length | 253 |
| Max. P | 0.999772 |

| Location | 21,348,202 – 21,348,303 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 79.30 |
| Mean single sequence MFE | -17.40 |
| Consensus MFE | -12.60 |
| Energy contribution | -12.72 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.759798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21348202 101 + 22407834 UAUUUAAGUGAAUUACCCAUUAACGAAACUUCCUUAAGGCUAACGG-------CAAUUGGGCAAAAGCUGCCUUGCGGCACAAAAAAUAUCACUAUAUUUUGAAUUUG ......(((((....((((((((.((....)).))))(.(....).-------)...)))).....(((((...)))))..........))))).............. ( -18.60) >DroSec_CAF1 58000 85 + 1 CAAUUAAGUGAAUUAC-CAUAAACGAAACUUCCUUAAGGCUAA--------------------AAAAAUGCCUUGAGGCACGAAAAAUAUCAAUAUAUUUUGAAUU-- .......(((......-(......)......(((((((((...--------------------......))))))))))))........((((......))))...-- ( -15.60) >DroSim_CAF1 44475 105 + 1 CAAUUAAGUGAAUUAC-CAUAAACAAAACUUCCUUAAGGCUAACGGCUCUUAGCAAUUAGGCAAAAAAUGCCUUGAGGCACGAAAAAUAUCAAUAUAUUUUGAAUU-- .......(((......-)))...........(((((((((.....((.((........)))).......)))))))))...........((((......))))...-- ( -18.00) >consensus CAAUUAAGUGAAUUAC_CAUAAACGAAACUUCCUUAAGGCUAACGG_______CAAUU_GGCAAAAAAUGCCUUGAGGCACGAAAAAUAUCAAUAUAUUUUGAAUU__ ...............................(((((((((.............................)))))))))..(((((.(((....))).)))))...... (-12.60 = -12.72 + 0.11)


| Location | 21,348,275 – 21,348,376 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 87.88 |
| Mean single sequence MFE | -21.33 |
| Consensus MFE | -18.06 |
| Energy contribution | -18.07 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21348275 101 - 22407834 AGAAGCGAAAGUUUCCCAGAAGAGUAGGAGAUAAGGGCUCCCAAGCUUCAAACCUAAAACCGAAAUUCAAAUUCAAAUUCAAAAUAUAGUGAUAUUUUUUG .(((((....)))))...((((..(.((((.......)))).)..))))..............................(((((.(((....))).))))) ( -19.70) >DroSec_CAF1 58059 95 - 1 AGAAGCGAAAGCUUCCUAGAGGAGUAGGAGCUUAGGGCUCACAAGCGACAAACCUAAAACCGAAAUUUU------AAUUCAAAAUAUAUUGAUAUUUUUCG .(((((....)))))...((((((((.((((.....)))).............................------...((((......)))))))))))). ( -21.80) >DroSim_CAF1 44554 95 - 1 AGAAGCGAAAGUUUCCUAGAGGAGUAGGAGCUUAGGGCUCCCAAGCGACAAACCUAAAACCGAAAUUUU------AAUUCAAAAUAUAUUGAUAUUUUUCG .(((((....)))))...(((((((((..((((.((....))))))..)....................------...((((......)))))))))))). ( -22.50) >consensus AGAAGCGAAAGUUUCCUAGAGGAGUAGGAGCUUAGGGCUCCCAAGCGACAAACCUAAAACCGAAAUUUU______AAUUCAAAAUAUAUUGAUAUUUUUCG .(((((....)))))...((((((((((...((((((((....)))......)))))..)).................((((......)))))))))))). (-18.06 = -18.07 + 0.01)

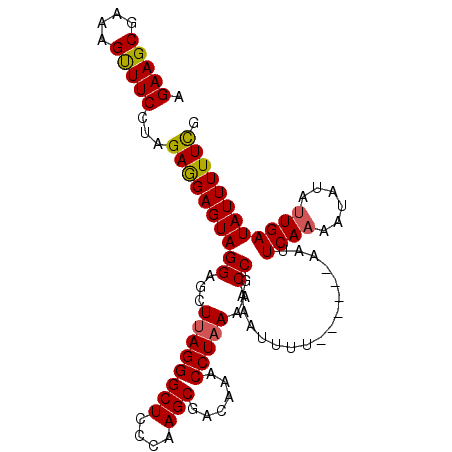
| Location | 21,348,336 – 21,348,455 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.77 |
| Mean single sequence MFE | -35.67 |
| Consensus MFE | -19.04 |
| Energy contribution | -20.27 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.53 |
| SVM decision value | 4.05 |
| SVM RNA-class probability | 0.999772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21348336 119 - 22407834 AAUAGAGGAUACGACGUUCAUCUUUUAUAGGUGAAGGUUGUGCUC-UUCAAAAAACUGGAAGCGCUUGGUCUAGUUCCCAAGAAGCGAAAGUUUCCCAGAAGAGUAGGAGAUAAGGGCUC ....(((((((((((.(((((((.....))))))).)))))).))-)))...........(((.((((.((((.((..(..(((((....)))))...)..)).))))...)))).))). ( -39.80) >DroSec_CAF1 58114 93 - 1 ---------------------A-UUUAUGUAUG-----UAUGCACGUGCAUAAACCCGGACCCAUUUGGUCUAGUUCCCAAGAAGCGAAAGCUUCCUAGAGGAGUAGGAGCUUAGGGCUC ---------------------.-((((((((((-----(....)))))))))))((((((((.....)))))((((((...(((((....)))))((.....))..))))))..)))... ( -34.80) >DroSim_CAF1 44609 93 - 1 ---------------------A-UUUAUGUAUG-----UAUGCACGUGCAUAAACCCGGACCCGUUUGGUCUAGUUCCCAAGAAGCGAAAGUUUCCUAGAGGAGUAGGAGCUUAGGGCUC ---------------------.-((((((((((-----(....)))))))))))((((((((.....)))))((((((...(((((....)))))((.....))..))))))..)))... ( -32.40) >consensus _____________________A_UUUAUGUAUG_____UAUGCACGUGCAUAAACCCGGACCCGUUUGGUCUAGUUCCCAAGAAGCGAAAGUUUCCUAGAGGAGUAGGAGCUUAGGGCUC .......................((((((((((...........))))))))))((((((((.....)))))((((((...(((((....)))))(.....)....))))))..)))... (-19.04 = -20.27 + 1.23)

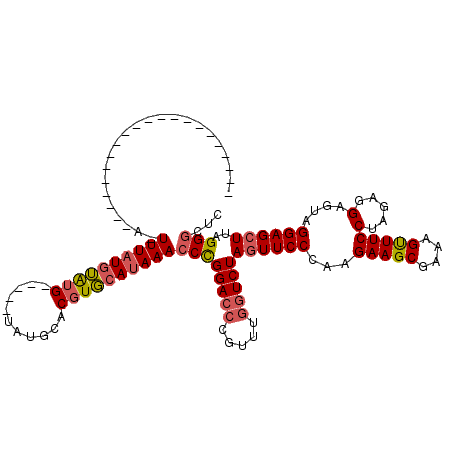
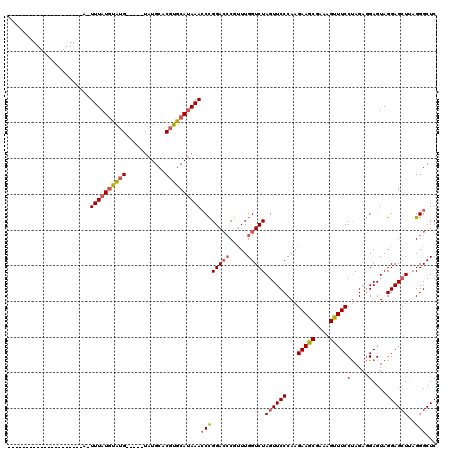
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:12 2006