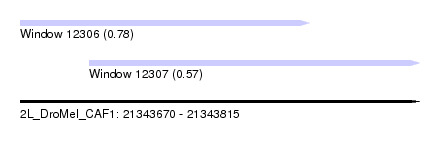
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,343,670 – 21,343,815 |
| Length | 145 |
| Max. P | 0.780071 |

| Location | 21,343,670 – 21,343,775 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 80.22 |
| Mean single sequence MFE | -23.50 |
| Consensus MFE | -17.16 |
| Energy contribution | -15.80 |
| Covariance contribution | -1.36 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.780071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21343670 105 + 22407834 CAUUAUAUUUCUCUUAUCUAAC-AAAGCUCACUGGGCUUCUUAUUUUUCUGGUGGGAGGCAUGGUCCAACUAAACUAUGCACAUUAUUCGAACUUCGUUAGCGAUC .................(((((-.((((((((..((...........))..)))))..(((((((........)))))))............))).)))))..... ( -21.90) >DroPse_CAF1 29424 94 + 1 ------------UUAAUCUCGCUAUAGCUCACUGGACUGCUCAUCUUUUUGGUGGGUGGCAUGGUCCAAUUGAAUUAUGCCCACUAUUCGAACUUUGUUAGUGAUC ------------........((....))(((((((...(((((((.....)))))))((((((((.(....).))))))))................))))))).. ( -24.00) >DroEre_CAF1 36926 103 + 1 CAUUGUAUG--UCUUAUCUAAC-AAAGCUCACUGGACUUCUUAUCUUUCUGGUGGGAGGCAUGGUCCAACUAAACUAUGCACACUAUUCGAACUUCGUUAGCGAUC .........--......(((((-.((((((((..((...........))..)))))..(((((((........)))))))............))).)))))..... ( -22.80) >DroYak_CAF1 41656 105 + 1 CAUUAUAUGUAUCUUAUCUAAC-AAAGCUCACUGGACUUCUUAUUUUUCUGGUGGGAGGCAUGGUCCAACUAAACUAUGCACACUAUUCGAACUUCGUUAGCGAUC .................(((((-.((((((((..((...........))..)))))..(((((((........)))))))............))).)))))..... ( -22.80) >DroAna_CAF1 27421 102 + 1 G---CUAUUUAUUUUAUCUACU-UUAGCUAACUGGACUACUCAUCUUUUUGGUGGGUGGAAUGGUCCAAUUGAAUUAUGCACAUUAUUCAAACUUUGUUAGUGACC .---..................-...(((((((((((((((((((.....))))))).....)))))).((((((.((....)).)))))).....)))))).... ( -23.70) >DroPer_CAF1 30622 94 + 1 ------------UUAAUCUCGCUAUAGCUCACUGGACUGCUCAUCUUUCUGGUGGGUGGCAUGGUCCAAUUGAAUUAUGCCCACUAUUCGAACUUUGUUAGUGAUC ------------......((((((..((((((..((..(.....)..))..))))))((((((((.(....).)))))))).................)))))).. ( -25.80) >consensus C____UAU___UCUUAUCUAAC_AAAGCUCACUGGACUUCUCAUCUUUCUGGUGGGAGGCAUGGUCCAACUAAACUAUGCACACUAUUCGAACUUCGUUAGCGAUC ..........................(((.((.((..((((((((.....))))))))(((((((........)))))))............))..)).))).... (-17.16 = -15.80 + -1.36)

| Location | 21,343,695 – 21,343,815 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.28 |
| Mean single sequence MFE | -38.43 |
| Consensus MFE | -22.41 |
| Energy contribution | -22.63 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568203 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
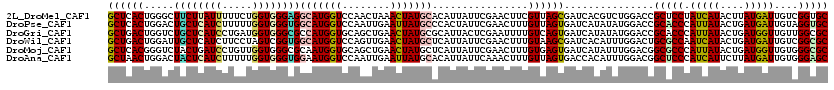
>2L_DroMel_CAF1 21343695 120 + 22407834 GCUCACUGGGCUUCUUAUUUUUCUGGUGGGAGGCAUGGUCCAACUAAACUAUGCACAUUAUUCGAACUUCGUUAGCGAUCACGUCUGGACCGCUCCUAUCAUACUUAUGAUUGUCGGUGC ...((((((.(...((((.....((((((((((((((((........)))))))......((((.((.(((....)))....)).))))...))))))))).....))))..))))))). ( -30.50) >DroPse_CAF1 29438 120 + 1 GCUCACUGGACUGCUCAUCUUUUUGGUGGGUGGCAUGGUCCAAUUGAAUUAUGCCCACUAUUCGAACUUUGUUAGUGAUCAUAUAUGGACCGCACCCAUUAUACUGAUGAUUGUAGGUGC ...((((..((...(((((....(((((((((...(((((((.......((((..(((((..(((...))).)))))..))))..))))))))))))))))....)))))..)).)))). ( -38.30) >DroGri_CAF1 39249 120 + 1 GCUGACUGGUCUGCUCAUCCUGAUGGUGGGCGCCAUGGUGCAGCUGAACUAUGCGCAUUACUCGAAUUUUGUCAGUGAUCAUAUAUGGACCGCACCCAUUAUACUGAUGGUUGUUGGCGC ((..((.((((((((((((.....)))))))((((((((........)))))).))((((((.((......)))))))).......)))))....(((((.....)))))..))..)).. ( -39.40) >DroWil_CAF1 30408 120 + 1 GCUGACUGGAUUGCUCAUCUUCCUAGUCGGUGGCAUGGUCCAGUUGAACUAUGCUCAUUAUUCGAACUUUGUAAGCGAUCACAUUUGGACUGCGCCAAUCAUACUGAUGAUUGUCGGCGC .(..(.(((((((((.((......(((((((((((((((.(....).))))))).)))....)).)))..)).))))))).)).)..)...(((((((((((....))))))...))))) ( -33.70) >DroMoj_CAF1 41129 120 + 1 GCUCACGGGUCUACUGAUCCUGUUGGUGGGCGCAAUGGUGCAGCUGAACUAUGCUCAUUAUUCGAACUUUGUGAGUGAUCAUAUUUGGACGGCGCCCAUUAUACUGAUGGUUGUGGGCGC (((((((((((....))))).(((((((((((((((((((.(((........))))))))))((..(..(((((....)))))...)..)))))))))))).))........)))))).. ( -49.00) >DroAna_CAF1 27443 120 + 1 GCUAACUGGACUACUCAUCUUUUUGGUGGGUGGAAUGGUCCAAUUGAAUUAUGCACAUUAUUCAAACUUUGUUAGUGACCACAUUUGGACGGCUCCCAUCAUUCUUAUGAUUGUGGGAGC (((((((((((((((((((.....))))))).....)))))).((((((.((....)).)))))).....))))))..(((....)))...(((((((.((.((....)).))))))))) ( -39.70) >consensus GCUCACUGGACUGCUCAUCUUUUUGGUGGGUGGCAUGGUCCAACUGAACUAUGCACAUUAUUCGAACUUUGUUAGUGAUCACAUUUGGACCGCACCCAUCAUACUGAUGAUUGUCGGCGC ((((((.....((((((((.....))))))))(((((((........)))))))................))))))...............(((((.(((((....)))))....))))) (-22.41 = -22.63 + 0.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:42:10 2006