| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,261,911 – 21,262,070 |
| Length | 159 |
| Max. P | 0.837821 |

| Location | 21,261,911 – 21,262,031 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.17 |
| Mean single sequence MFE | -50.10 |
| Consensus MFE | -35.73 |
| Energy contribution | -35.27 |
| Covariance contribution | -0.46 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
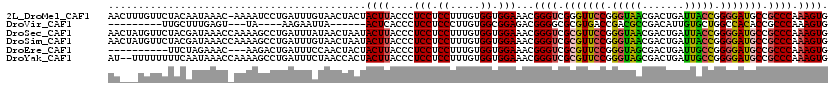
>2L_DroMel_CAF1 21261911 120 - 22407834 ACUACUUACCCUCCUCCUUUGUGGUGGAAACGGGUCGGGUUCCGGGUAACGACUGAUUACCGGGGAUGCCGCCCAAAGUGACUCCACUACCGCCCAAGGAAACGGAUGCGCCACCGCUUC ...........((((((((.(.(((((....(((.((((((((.(((((.......))))).))))).))))))..((((....)))).)))))))))))...))).(((....)))... ( -43.50) >DroVir_CAF1 153088 117 - 1 ---ACUCACCCUCCUCCCUUGUGGCGGAGACGGGGCGCGUGACCGACGCCGACAUUGUGCUGGCCACACCGCCCAAAGUGGCGCCACUGCCGCCCAGAGGAAUGGCUGUGCCGCCACUGC ---.......((((.((.....)).))))...(((((.(((.(((.(((.......))).))).)))..)))))..(((((((.(((.(((.((....))...))).))).))))))).. ( -49.20) >DroPse_CAF1 802 120 - 1 AGCUCUUACCCUCCUCCUUUGUGGAUGAAACGGGUCGCGUUCCCGGCAGCGACUGGUUGCUGGAGACGCCUCCCAGUGUGGCACCACUGCCGCCCAGGGGAAUGGCAUUGCCUCCGCUCC (((.....(((((.(((.....))).))...(((..((((..((((((((.....))))))))..))))..))).(.((((((....)))))).).)))....(((...)))...))).. ( -50.60) >DroGri_CAF1 47951 120 - 1 GACUCUUACCCUCCUCCUUUGUGCUGGAGGCGGGGCGUGUGCCCGGCGCAGUCAUUGGGAUGGCAACGCCGCCCAAAGUGGCGCCACUGCCGCCCAGGGGAAUGGCGGAGCCACCACUGC ((((....(((.(((((........))))).)))((((.......))))))))....((.((((..((((.(((...((((((....))))))....)))...))))..))))))..... ( -55.60) >DroEre_CAF1 688 120 - 1 ACUACUUACCCUCCUCCUUUGUGGUGGAAACGGGUCGCGUUCCGGGUAGCGACUGAUUGCCGGGGAUGCCGCCCAAAGUGGCUCCACUACCGCCCAAGGGAACGGAUGCACCACCGCUUC ...........((((((((((.(((((....((((.(((((((.(((((.......))))).))))))).))))..((((....)))).))))))))))))..))).((......))... ( -51.20) >DroPer_CAF1 802 120 - 1 AGCUCUUACCCUCCUCCUUUGUGGAUGAAACGGGUCGCGUUCCCGGCAGCGACUGGUUGCUGGAGACGCCUCCCAGUGUGGCGCCACUGCCGCCCAGGGGAAUGGCAUUGCCUCCGCUAC (((.....(((((.(((.....))).))...(((..((((..((((((((.....))))))))..))))..))).(.((((((....)))))).).)))....(((...)))...))).. ( -50.50) >consensus ACCACUUACCCUCCUCCUUUGUGGUGGAAACGGGUCGCGUUCCCGGCAGCGACUGAUUGCUGGAGACGCCGCCCAAAGUGGCGCCACUGCCGCCCAGGGGAAUGGCUGUGCCACCGCUGC ....................(((((((....(((..(((((.(((((((.......))))))).)))))..)))...(((....))).((((((....))..))))....)))))))... (-35.73 = -35.27 + -0.46)

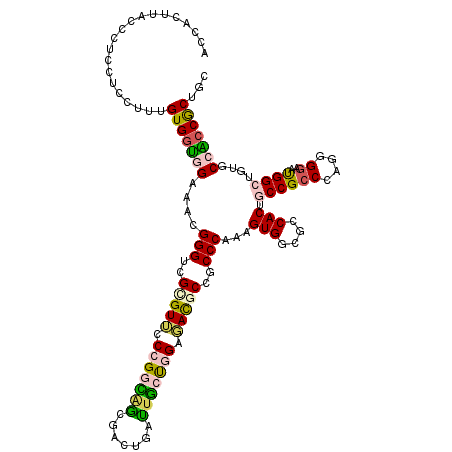
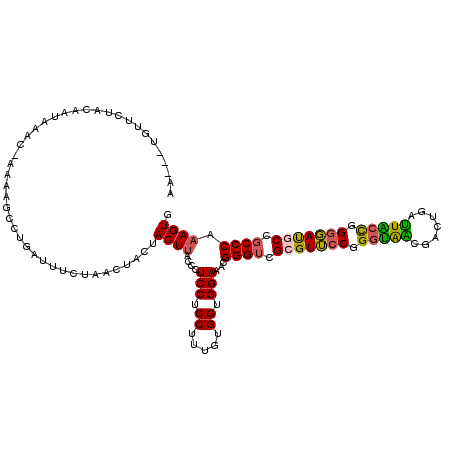
| Location | 21,261,951 – 21,262,070 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.33 |
| Mean single sequence MFE | -34.60 |
| Consensus MFE | -29.54 |
| Energy contribution | -29.02 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837821 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21261951 119 - 22407834 AACUUUGUUCUACAAUAAAC-AAAAUCCUGAUUUGUAACUACUACUUACCCUCCUCCUUUGUGGUGGAAACGGGUCGGGUUCCGGGUAACGACUGAUUACCGGGGAUGCCGCCCAAAGUG .((((((..(..((......-.....((((((((((..(((((((...............)))))))..))))))))))((((.(((((.......))))).))))))..)..)))))). ( -34.76) >DroVir_CAF1 153128 98 - 1 ---------UUGCUUUGAGU---UA----AAGAAUUA------ACUCACCCUCCUCCCUUGUGGCGGAGACGGGGCGCGUGACCGACGCCGACAUUGUGCUGGCCACACCGCCCAAAGUG ---------..(((((((((---((----(....)))------))))...((((.((.....)).))))...(((((.(((.(((.(((.......))).))).)))..)))))))))). ( -34.40) >DroSec_CAF1 821 120 - 1 AACUAUGUUCUACGAUAAACCAAAAGCCUGAUUUAUAACUAAUACUUACCCUCCUCCUUUGUGGUGGAAACGGGUCGCGUUCCGGGUAACGACUGAUUACCGGGGAUGCCGCCCAAAGUG ..............(((((.((......)).)))))......(((((....(((.((.....)).)))...((((.(((((((.(((((.......))))).))))))).)))).))))) ( -34.50) >DroSim_CAF1 734 120 - 1 AACUAUGUUCUACGAUAAACCAAAAGCCUGAUUUGUAACUAAUACUUACCCUCCUCCUUUGUGGUGGAAACGGGUCGCGUUCCGGGUAACGACUGAUUACCGGGGAUGCCGCCCAAAGUG ..............(((((.((......)).)))))......(((((....(((.((.....)).)))...((((.(((((((.(((((.......))))).))))))).)))).))))) ( -34.50) >DroEre_CAF1 728 107 - 1 ----------UUCUAGAAAC---AAGACUGAUUUCCAACUACUACUUACCCUCCUCCUUUGUGGUGGAAACGGGUCGCGUUCCGGGUAGCGACUGAUUGCCGGGGAUGCCGCCCAAAGUG ----------..........---...(((..((((((.((((..................)))))))))).((((.(((((((.(((((.......))))).))))))).))))..))). ( -34.57) >DroYak_CAF1 729 118 - 1 AU--UUUUUUUUCAAUAAACCAAAAGCCUGAUUUCUAACCACUACUUACCCUCCUCCUUUGUGGUGGAAACGGGUCGCGUUCCGGGUAGCGACUGAUUGCCGGGGAUGCCGCCCAAAGUG ..--..................................(((((((...............)))))))....((((.(((((((.(((((.......))))).))))))).))))...... ( -34.86) >consensus AA___UGUUCUACAAUAAAC_AAAAGCCUGAUUUCUAACUACUACUUACCCUCCUCCUUUGUGGUGGAAACGGGUCGCGUUCCGGGUAACGACUGAUUACCGGGGAUGCCGCCCAAAGUG ...........................................((((....(((.((.....)).)))...((((.(((((((.(((((.......))))).))))))).)))).)))). (-29.54 = -29.02 + -0.52)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:51 2006