| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,215,373 – 21,215,476 |
| Length | 103 |
| Max. P | 0.575550 |

| Location | 21,215,373 – 21,215,476 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 83.04 |
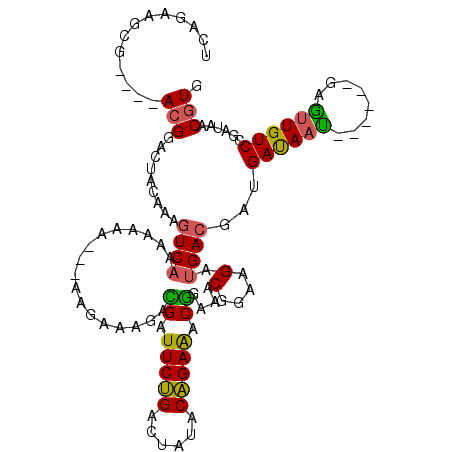
| Mean single sequence MFE | -20.80 |
| Consensus MFE | -14.75 |
| Energy contribution | -14.20 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
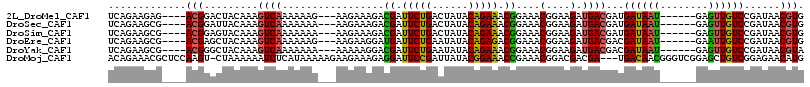
>2L_DroMel_CAF1 21215373 103 + 22407834 UCAGAAGAG----ACGGACUACAAAGUCAAAAAAG---AAGAAAGACGAUUCUGACUAUACAGAAACGGAAACGGAAGAUGACGAUGAUAAU------GAGUUGUCCGAUAACGUG .........----.(((((.((...((((......---...........(((((......))))).((....)).....)))).((....))------..)).)))))........ ( -20.20) >DroSec_CAF1 5505 103 + 1 UCAGAAGCG----ACGGAUUACAAAGUCAAAAAAA---AAGAAAGACGAUUCUGACUAUACAGAAACGGAAACGGAAGAUGACGAUGAUAAU------GAGUUGUCCGAUAACGUG ......(((----((..((((((..((((......---...........(((((......))))).((....)).....))))..)).))))------..)))))........... ( -20.20) >DroSim_CAF1 5573 103 + 1 UCAGAAGCG----ACGGAGUACAAAGUCAAAAAAA---AAGAAAGACGAUUCUGACUAUACAGAAACGGAAACGGAAGAUGACGAUGAUAAU------GAGUUGUCCGAUAACGUG ......(((----((....((((..((((......---...........(((((......))))).((....)).....))))..)).))..------..)))))........... ( -17.80) >DroEre_CAF1 5719 103 + 1 UCAGAAGCG----ACGAGCUACAAAGUCAAAAAAG---AAGAAGGAUGAUUCUGAAUAUACAGAGACGGAAACGGAAGAUGACGACGAUAAU------GAAUUGUCCGAUAACGUG ...(.(((.----....))).)...((((......---...........(((((......))))).((....)).....))))((((((...------..)))))).......... ( -19.40) >DroYak_CAF1 5534 103 + 1 UCAGAAGCG----ACGGGCUACAAAGUCAAAAAAA---AAAAAGGACGAUUCUGAAUAUACAGAAACGGAAACGGAAGAUGACGACGAUAAU------GAGUUGUCCGAUAACGUA ......(((----.(((((......((((......---...........(((((......))))).((....)).....))))(((......------..))))))))....))). ( -21.40) >DroMoj_CAF1 6415 112 + 1 ACAGAAACGCUCCAAGU-CUAAAAAAUCUCAUAAAAAGAAGAAAGAGGAUUCCGAUUAUACGGAAACCGAAACGGACGACGA---UGACAACGGGUCGGAGCUGUCGGAGAACAUG ...((...(((((..((-((......(((.......))).......((.(((((......))))).)).....)))).....---.(((.....))))))))..)).......... ( -25.80) >consensus UCAGAAGCG____ACGGACUACAAAGUCAAAAAAA___AAGAAAGACGAUUCUGACUAUACAGAAACGGAAACGGAAGAUGACGAUGAUAAU______GAGUUGUCCGAUAACGUG .............(((.........((((.................((.(((((......))))).))....(....).))))...((((((........))))))......))). (-14.75 = -14.20 + -0.55)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:39 2006