| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,213,744 – 21,213,880 |
| Length | 136 |
| Max. P | 0.993882 |

| Location | 21,213,744 – 21,213,840 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 77.40 |
| Mean single sequence MFE | -31.48 |
| Consensus MFE | -15.50 |
| Energy contribution | -16.17 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.626638 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21213744 96 + 22407834 AUC---CGA---GGCCAGCGAGGCGGAGGAGGAGGCACGUAAUAAGAAGAAAAAAAAGUGUCCUGGCAAGGAUGGCUGGUCUUCCGAUUCAGAAGAACAGCC (((---(..---.(((((.((.((.................................)).)))))))..))))(((((.(((((.......))))).))))) ( -29.71) >DroVir_CAF1 4393 99 + 1 GUCCUUUGAGUCGGCCAGCGAGGCCGAAG---AGACCAAAAAGAAGAUCAAGAAAAAAUGCCCCGGCAAGGAUGGCUGGUCCUCGGACUCCGAGGAGCAGGC (((((((((.((((((.....))))))..---...............)))).......(((....))))))))(.(((.(((((((...))))))).))).) ( -36.87) >DroGri_CAF1 4765 99 + 1 GUCCUUUGAGUCAGCCAGCGAAGCGGAAG---AGGCCAAAAAGAAGAUCAAGAAGAAAUGUCCAGGCAAAGAUGGCUGGUCCUCAGACUCUGAGGAACAGGC .(((((.(((((.((.......))(((..---.(((((.........((.....))..(((....)))....)))))..)))...))))).)))))...... ( -28.10) >DroEre_CAF1 3742 96 + 1 GUC---GGA---GGCCAGCGAGGCGGAGGAGGAGACCCGUAAUAAGAAGAAAAAGAAGUGUCCUGGUAAGGAUGGCUGGUCCUCCGAUUCAGAAGAGCAGCC (((---(((---(((((((...((((....(....)))))...................(((((....))))).))))).)))))))).............. ( -34.90) >DroYak_CAF1 3655 96 + 1 AUC---UGA---GGCUAGCGAGGCGGAGGAGGAGGCCCGUAAUAAGAAGAAAAAAAAGUGUCCUGGCAAGGAUGGCUGGUCCUCCGAUUCAGAGGAACAGCC .((---(((---((((.....)))((((((...(((((.......).............(((((....)))))))))..))))))..))))))......... ( -29.30) >DroAna_CAF1 3873 96 + 1 GUC---CGA---GGCCAGUGAAGCCGAGGAGGAGGCUCGCAGUAAGAAGAAGAAGAAGUGUCCCGGAAAGGACGGUUGGUCGUCCGACUCCGAGGAGCAACC .((---(..---(((.......)))..)))((..(((((((.(.............).)))..((((..((((((....))))))...))))..))))..)) ( -30.02) >consensus GUC___UGA___GGCCAGCGAGGCGGAGGAGGAGGCCCGUAAUAAGAAGAAAAAAAAGUGUCCUGGCAAGGAUGGCUGGUCCUCCGACUCAGAGGAACAGCC .............(((.....)))...................................((((......))))(((((.(((((.......))))).))))) (-15.50 = -16.17 + 0.67)

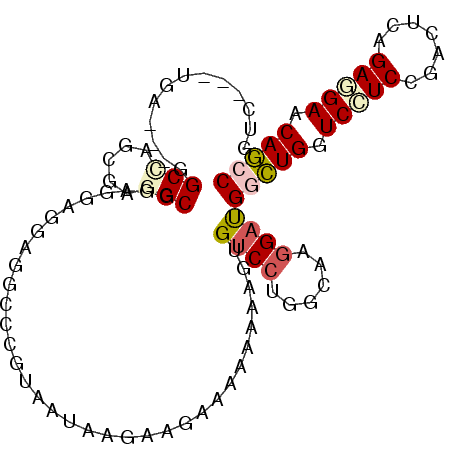

| Location | 21,213,744 – 21,213,840 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 77.40 |
| Mean single sequence MFE | -29.56 |
| Consensus MFE | -19.92 |
| Energy contribution | -19.37 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21213744 96 - 22407834 GGCUGUUCUUCUGAAUCGGAAGACCAGCCAUCCUUGCCAGGACACUUUUUUUUCUUCUUAUUACGUGCCUCCUCCUCCGCCUCGCUGGCC---UCG---GAU (((((.(((((((...))))))).)))))((((.....(((.(((...................))))))........(((.....))).---..)---))) ( -28.21) >DroVir_CAF1 4393 99 - 1 GCCUGCUCCUCGGAGUCCGAGGACCAGCCAUCCUUGCCGGGGCAUUUUUUCUUGAUCUUCUUUUUGGUCU---CUUCGGCCUCGCUGGCCGACUCAAAGGAC (.(((.(((((((...))))))).))).).(((((...(((............((((........)))).---..((((((.....))))))))).))))). ( -36.40) >DroGri_CAF1 4765 99 - 1 GCCUGUUCCUCAGAGUCUGAGGACCAGCCAUCUUUGCCUGGACAUUUCUUCUUGAUCUUCUUUUUGGCCU---CUUCCGCUUCGCUGGCUGACUCAAAGGAC ......((((..(((((.(((((((((.((....)).)))).((........)).))))).....((((.---.............)))))))))..)))). ( -26.34) >DroEre_CAF1 3742 96 - 1 GGCUGCUCUUCUGAAUCGGAGGACCAGCCAUCCUUACCAGGACACUUCUUUUUCUUCUUAUUACGGGUCUCCUCCUCCGCCUCGCUGGCC---UCC---GAC ...............(((((((.(((((..((((....)))).....................((((........))))....)))))))---)))---)). ( -28.60) >DroYak_CAF1 3655 96 - 1 GGCUGUUCCUCUGAAUCGGAGGACCAGCCAUCCUUGCCAGGACACUUUUUUUUCUUCUUAUUACGGGCCUCCUCCUCCGCCUCGCUAGCC---UCA---GAU (((((.(((((((...))))))).))))).((((....))))..........(((........(((((..........))).))......---..)---)). ( -26.19) >DroAna_CAF1 3873 96 - 1 GGUUGCUCCUCGGAGUCGGACGACCAACCGUCCUUUCCGGGACACUUCUUCUUCUUCUUACUGCGAGCCUCCUCCUCGGCUUCACUGGCC---UCG---GAC ((..(((((((((((..(((((......))))).)))))))...............(.....).))))..))(((..((((.....))))---..)---)). ( -31.60) >consensus GGCUGCUCCUCUGAAUCGGAGGACCAGCCAUCCUUGCCAGGACACUUCUUCUUCUUCUUAUUACGGGCCUCCUCCUCCGCCUCGCUGGCC___UCA___GAC (((((.(((((((...))))))).))))).((((....))))....................................(((.....)))............. (-19.92 = -19.37 + -0.55)

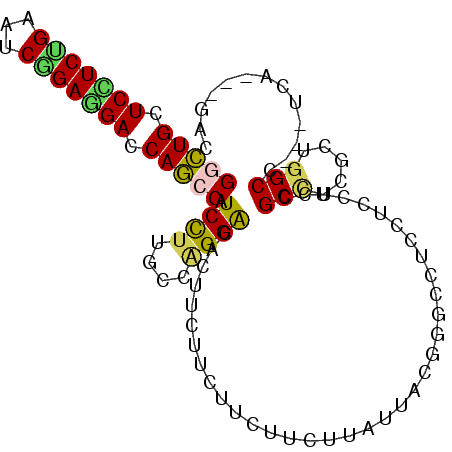

| Location | 21,213,778 – 21,213,880 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 84.44 |
| Mean single sequence MFE | -29.82 |
| Consensus MFE | -19.22 |
| Energy contribution | -19.42 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.970925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21213778 102 + 22407834 AAUAAGAAG------------AAAAAAAAGUGUCCUGGCAAGGAUGG---CUGGUCUUCCGAUUCAGAAGAACAGCCAGAAAGUGAGGAGGAGGAGGAGGAGCCACCGCACUAUGAA .........------------.......((((((((.((.....(((---(((.(((((.......))))).))))))....)).))))...((.((.....)).)).))))..... ( -30.40) >DroSec_CAF1 3737 102 + 1 AAUAAGAAG------------AAAAAAAAGUGUCCUGGCAAGGAUGG---CUGGUCUUCUGAUUCAGAAGAACAGCCGGAAAGCGAGGAGGAAGAGGAGGAGCCACCGCACUAUGAA .........------------.......((((((((.((.....(((---(((.(((((((...))))))).))))))....)).))))......((.(....).)).))))..... ( -32.90) >DroEre_CAF1 3776 102 + 1 AAUAAGAAG------------AAAAAGAAGUGUCCUGGUAAGGAUGG---CUGGUCCUCCGAUUCAGAAGAGCAGCCAGAGAGCGAGGAGGAAGAGGAGGAACCACCGCACUAUGAA .........------------.......((((((((....)))..((---.(((((((((..(((......((.........))......)))..))))).)))))))))))..... ( -29.30) >DroWil_CAF1 4314 117 + 1 AAUCAGAUGAAAAAGAAGGCGAAAAAGAAAUGUCCUGGCAAAGAUGGAACCUGGUCAUCAGACUCCGAAGAGCAAGCUGAGAGUGAGGAGGAAGAAGAGGAGCCACCUCAUUAUGAA ..(((........((...((..........(((....)))....((((..(((.....)))..))))....))...))...(((((((.((...........)).))))))).))). ( -21.80) >DroYak_CAF1 3689 102 + 1 AAUAAGAAG------------AAAAAAAAGUGUCCUGGCAAGGAUGG---CUGGUCCUCCGAUUCAGAGGAACAGCCAGAGAGCGAGGAGGAAGAGGAGGAGCCACCUCACUAUGAA .........------------.......(((.((((.((.....(((---(((.(((((.......))))).))))))....)).))))....((((.(....).)))))))..... ( -34.70) >consensus AAUAAGAAG____________AAAAAAAAGUGUCCUGGCAAGGAUGG___CUGGUCCUCCGAUUCAGAAGAACAGCCAGAGAGCGAGGAGGAAGAGGAGGAGCCACCGCACUAUGAA ............................((((((((.((.....(((...(((.(((((.......))))).))))))....)).))))......((.(....).)).))))..... (-19.22 = -19.42 + 0.20)

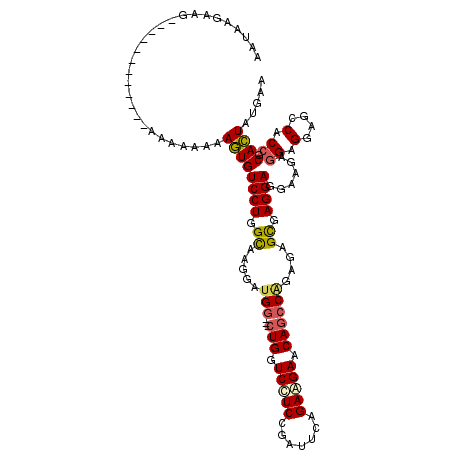
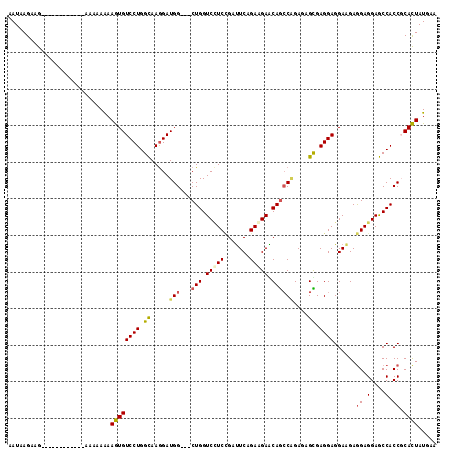
| Location | 21,213,778 – 21,213,880 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 84.44 |
| Mean single sequence MFE | -28.17 |
| Consensus MFE | -18.08 |
| Energy contribution | -17.40 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.29 |
| Mean z-score | -4.07 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.993882 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21213778 102 - 22407834 UUCAUAGUGCGGUGGCUCCUCCUCCUCCUCCUCACUUUCUGGCUGUUCUUCUGAAUCGGAAGACCAG---CCAUCCUUGCCAGGACACUUUUUUUU------------CUUCUUAUU .....((((.((.((...........)).)).))))...((((((.(((((((...))))))).)))---)))((((....))))...........------------......... ( -27.30) >DroSec_CAF1 3737 102 - 1 UUCAUAGUGCGGUGGCUCCUCCUCUUCCUCCUCGCUUUCCGGCUGUUCUUCUGAAUCAGAAGACCAG---CCAUCCUUGCCAGGACACUUUUUUUU------------CUUCUUAUU .....((((.((.(....).))......((((.((.....(((((.(((((((...))))))).)))---))......)).)))))))).......------------......... ( -28.80) >DroEre_CAF1 3776 102 - 1 UUCAUAGUGCGGUGGUUCCUCCUCUUCCUCCUCGCUCUCUGGCUGCUCUUCUGAAUCGGAGGACCAG---CCAUCCUUACCAGGACACUUCUUUUU------------CUUCUUAUU .....((((.((.((...........)).)).))))...((((((.(((((((...))))))).)))---)))((((....))))...........------------......... ( -27.60) >DroWil_CAF1 4314 117 - 1 UUCAUAAUGAGGUGGCUCCUCUUCUUCCUCCUCACUCUCAGCUUGCUCUUCGGAGUCUGAUGACCAGGUUCCAUCUUUGCCAGGACAUUUCUUUUUCGCCUUCUUUUUCAUCUGAUU ........((((((((..........(((..(((...((((...((((....)))))))))))..)))..........)))((((....))))....)))))............... ( -23.35) >DroYak_CAF1 3689 102 - 1 UUCAUAGUGAGGUGGCUCCUCCUCUUCCUCCUCGCUCUCUGGCUGUUCCUCUGAAUCGGAGGACCAG---CCAUCCUUGCCAGGACACUUUUUUUU------------CUUCUUAUU .....(((((((.((...........)).)))))))...((((((.(((((((...))))))).)))---)))((((....))))...........------------......... ( -33.80) >consensus UUCAUAGUGCGGUGGCUCCUCCUCUUCCUCCUCGCUCUCUGGCUGUUCUUCUGAAUCGGAAGACCAG___CCAUCCUUGCCAGGACACUUUUUUUU____________CUUCUUAUU .....((((.((.((...........)).)).))))....(((((.(((((((...))))))).)))...)).((((....))))................................ (-18.08 = -17.40 + -0.68)

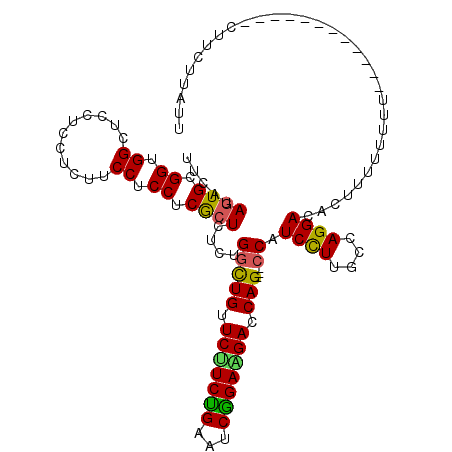

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:36 2006