| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,207,553 – 21,207,684 |
| Length | 131 |
| Max. P | 0.903501 |

| Location | 21,207,553 – 21,207,652 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 72.48 |
| Mean single sequence MFE | -22.87 |
| Consensus MFE | -9.72 |
| Energy contribution | -9.92 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.43 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21207553 99 + 22407834 CCAGCCCCCUCCC----GAAACGGGAAAGGCAUUCCAAAAUGU-U----AGAUGAAUG-CAUGCUAUUGUAAUUGGAACUGCUUGUUUGCAUGUUCUAAUAAACUACAC ..(((....((((----(...)))))...((((((........-.----....)))))-)..)))..(((((((((((((((......))).))))))))....)))). ( -26.12) >DroSec_CAF1 17874 99 + 1 CCAGCCCCCUCCC----GUAACGGGAAAGGCAUUCCUAAAUAU-U----AGAUGAAUG-CAUGUCAUUGUAAUUGGAACUGCUUGUUUGCAUGUUCUAAUAAACUACAC ...(((...((((----(...)))))..)))............-(----((.....((-((......))))(((((((((((......))).))))))))...)))... ( -25.30) >DroEre_CAF1 22544 94 + 1 CCAGCC--CUUCC----GUAACGGGAAAGGCAUUCCAAAAU--------AGAUGAAUG-CAUGUCAUUGUAAUUGGAACUGCUUGUUUGCAUGUUCUAAUAAACUACAC ...(((--.((((----......)))).))).........(--------((.....((-((......))))(((((((((((......))).))))))))...)))... ( -24.40) >DroYak_CAF1 21726 90 + 1 CCAGAC--CUCCC----GUAACGGGGAAAGCAUUCCAAAAU--------AGAU----G-CAUGUCAUUGUAAUUGGAACUGCUUGUUUGCAUGUUCUAAUAAACUACAC ...(((--.((((----......))))..(((((.......--------.)))----)-)..)))..(((((((((((((((......))).))))))))....)))). ( -24.90) >DroMoj_CAF1 20376 84 + 1 -------UUGUUUGUCAUUACCCA------UAUUUCAUAAU--------AAAUGUAUG-CAUGUCAUUCUUAUUAGAACUGGUUGUUUGCAUGUUCUAAUAAGCAA--- -------.....((.(((....((------(((........--------....)))))-.))).))..(((((((((((..((.....))..)))))))))))...--- ( -14.40) >DroAna_CAF1 14422 100 + 1 CACUGCCCCUCCA----CAAACUAUAA-----UUUCGUAAUAUGUAGGCAUGUGUAUGUCAUGUUAUUGUUAUUGGAACGGCUUGUUUGCAUGUUCUAAUAAACUACAU .............----..........-----.........(((((((((((.......))))).....(((((((((((((......)).))))))))))).)))))) ( -22.10) >consensus CCAGCC_CCUCCC____GUAACGGGAAAGGCAUUCCAAAAU________AGAUGAAUG_CAUGUCAUUGUAAUUGGAACUGCUUGUUUGCAUGUUCUAAUAAACUACAC .......................................................................(((((((((((......))).))))))))......... ( -9.72 = -9.92 + 0.20)
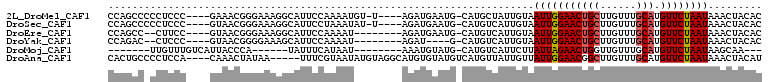
| Location | 21,207,578 – 21,207,684 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 77.85 |
| Mean single sequence MFE | -19.85 |
| Consensus MFE | -10.10 |
| Energy contribution | -10.43 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903501 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21207578 106 + 22407834 GCAUUCCAAAAUGUUAGAUGAAUGCAUGCUAUUGUAAUUGGAACUGCUUGUUUGCAUGUUCUAAUAAACUACACACACACACGACACGAAAAAUGACGCCAAUUCA ((((((.............)))))).......(((((((((((((((......))).))))))))....))))................................. ( -17.82) >DroPse_CAF1 14780 90 + 1 -UAUUUCAUAAU---------AUGCAUGUCGUUCUUAUUAGAACUGCUUGUUUGCAUGUUCCAAUAAACUACAAACACAAAUGC------AAAUUACGCCCGCACA -.......((((---------.(((((((.(((((....))))).)).((((((...(((......)))..))))))...))))------).)))).......... ( -16.60) >DroSec_CAF1 17899 106 + 1 GCAUUCCUAAAUAUUAGAUGAAUGCAUGUCAUUGUAAUUGGAACUGCUUGUUUGCAUGUUCUAAUAAACUACACACACACACGACACGAAAAAUGACGCCACUUCA (((((((((.....)))..))))))..((((((((((((((((((((......))).))))))))....))).........((...))...))))))......... ( -23.10) >DroEre_CAF1 22567 96 + 1 GCAUUCCAAAAU---AGAUGAAUGCAUGUCAUUGUAAUUGGAACUGCUUGUUUGCAUGUUCUAAUAAACUACACACAC-------ACGAGUAAUGACGCCAAUUCA ((((((......---....))))))..((((((...(((((((((((......))).))))))))..(((.(......-------..))))))))))......... ( -22.90) >DroYak_CAF1 21749 92 + 1 GCAUUCCAAAAU---AGAU----GCAUGUCAUUGUAAUUGGAACUGCUUGUUUGCAUGUUCUAAUAAACUACACACAC-------ACGAAUAAUGACGCCAAUUCA (((((.......---.)))----))..((((((((.(((((((((((......))).))))))))........(....-------..).))))))))......... ( -22.10) >DroPer_CAF1 14587 90 + 1 -UAUUUCAUAAU---------AUGCAUGUCGUUCUUAUUAGAACUGCUUGUUUGCAUGUUCCAAUAAACUACAAACACAAAUGC------AAAUUACGCCCGCACA -.......((((---------.(((((((.(((((....))))).)).((((((...(((......)))..))))))...))))------).)))).......... ( -16.60) >consensus GCAUUCCAAAAU___AGAU__AUGCAUGUCAUUGUAAUUGGAACUGCUUGUUUGCAUGUUCUAAUAAACUACACACACA_A_G__ACGAAAAAUGACGCCAAUUCA ....................................(((((((((((......))).))))))))......................................... (-10.10 = -10.43 + 0.33)
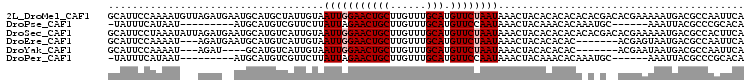
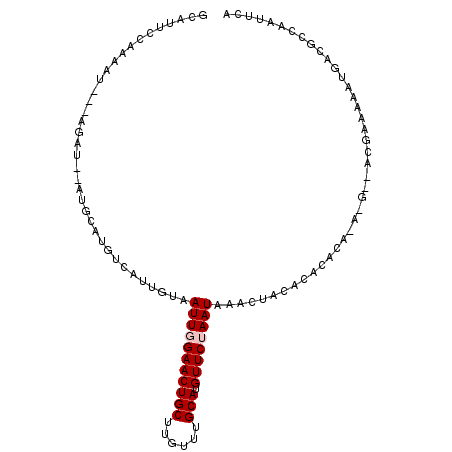

| Location | 21,207,578 – 21,207,684 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 77.85 |
| Mean single sequence MFE | -22.68 |
| Consensus MFE | -13.61 |
| Energy contribution | -13.92 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21207578 106 - 22407834 UGAAUUGGCGUCAUUUUUCGUGUCGUGUGUGUGUGUAGUUUAUUAGAACAUGCAAACAAGCAGUUCCAAUUACAAUAGCAUGCAUUCAUCUAACAUUUUGGAAUGC (((((..(((.(((.....))).)))((((((.(((((((.....((((.(((......))))))).)))))))...)))))))))))(((((....))))).... ( -24.90) >DroPse_CAF1 14780 90 - 1 UGUGCGGGCGUAAUUU------GCAUUUGUGUUUGUAGUUUAUUGGAACAUGCAAACAAGCAGUUCUAAUAAGAACGACAUGCAU---------AUUAUGAAAUA- ........((((((.(------((((...(((((.....((((((((((.(((......))))))))))))))))))..))))).---------)))))).....- ( -25.50) >DroSec_CAF1 17899 106 - 1 UGAAGUGGCGUCAUUUUUCGUGUCGUGUGUGUGUGUAGUUUAUUAGAACAUGCAAACAAGCAGUUCCAAUUACAAUGACAUGCAUUCAUCUAAUAUUUAGGAAUGC .......(((.(((.....))).)))((((...(((((((.....((((.(((......))))))).)))))))...))))((((((..(((.....))))))))) ( -22.40) >DroEre_CAF1 22567 96 - 1 UGAAUUGGCGUCAUUACUCGU-------GUGUGUGUAGUUUAUUAGAACAUGCAAACAAGCAGUUCCAAUUACAAUGACAUGCAUUCAUCU---AUUUUGGAAUGC .........((((((((....-------..)))(((((((.....((((.(((......))))))).))))))))))))..((((((....---......)))))) ( -19.70) >DroYak_CAF1 21749 92 - 1 UGAAUUGGCGUCAUUAUUCGU-------GUGUGUGUAGUUUAUUAGAACAUGCAAACAAGCAGUUCCAAUUACAAUGACAUGC----AUCU---AUUUUGGAAUGC .......((((((..((..((-------((((((((.((..(((.((((.(((......))))))).))).)).)).))))))----))..---))..))..)))) ( -18.10) >DroPer_CAF1 14587 90 - 1 UGUGCGGGCGUAAUUU------GCAUUUGUGUUUGUAGUUUAUUGGAACAUGCAAACAAGCAGUUCUAAUAAGAACGACAUGCAU---------AUUAUGAAAUA- ........((((((.(------((((...(((((.....((((((((((.(((......))))))))))))))))))..))))).---------)))))).....- ( -25.50) >consensus UGAAUUGGCGUCAUUUUUCGU__C_U_UGUGUGUGUAGUUUAUUAGAACAUGCAAACAAGCAGUUCCAAUUACAAUGACAUGCAU__AUCU___AUUUUGGAAUGC ............................((((((((.(((.(((.((((.(((......))))))).)))...))).))))))))..................... (-13.61 = -13.92 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:32 2006