| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,179,276 – 21,179,380 |
| Length | 104 |
| Max. P | 0.807087 |

| Location | 21,179,276 – 21,179,380 |
|---|---|
| Length | 104 |
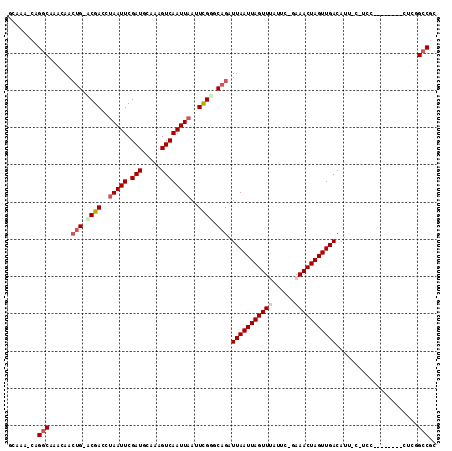
| Sequences | 6 |
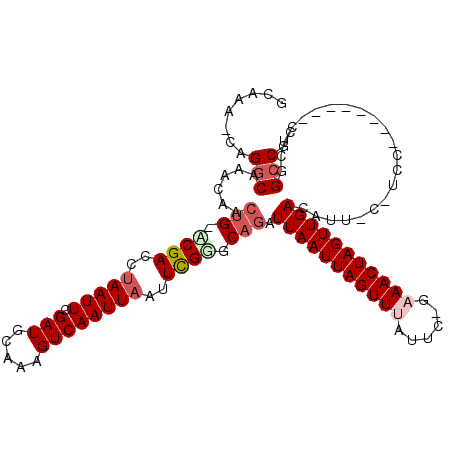
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 79.40 |
| Mean single sequence MFE | -27.57 |
| Consensus MFE | -19.08 |
| Energy contribution | -19.97 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807087 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21179276 104 - 22407834 GCAAA-CGGGCAAACAACUG-ACGACCCAAUUCGAUGCGUUGUCAAUUAAUUCGGGCAGAUUAAUUAGUUUAUGC-GAAACUAGUUGACAUUUCAUCC--------CGGUGCCGC .....-..((((......((-(((((.((......)).)))))))......(((((..(((((((((((((....-.)))))))))))....))..))--------))))))).. ( -29.30) >DroVir_CAF1 20692 98 - 1 ACAAACGAGGCAAACAACUGAACGACCUAAUUCGAUGCAAAGUCAAUUAAUUUGUGCAGAUUAAUUAGUUAAGUC-GAAACUAGUUGACAUC-----G-----------GGCCGC ........(((......(((.((((..(((((.(((.....))))))))..)))).))).((((((((((.....-..))))))))))....-----.-----------.))).. ( -21.10) >DroPse_CAF1 17686 112 - 1 GCCAA-CAGGCAAACAACUG-UCGACCUAAUUCGAUGCAAAGUCAAUUAAUUCGGGCAGAUUAAUUAGUUUGUUC-GAAACUAGUUGACAUUCCGUCCACACCGGGCUCGGCCGC (((..-..)))........(-(((((((.....((((.((.((((((((.((((((((((((....)))))))))-)))..)))))))).)).))))......))).)))))... ( -36.10) >DroMoj_CAF1 19941 101 - 1 AUGGGGCAGGCAAACAAGCGAACGACCUAAUUCGAUGCAAAGUCAAUUAAUUUGAGCAGAUUAAUUAGUUAAGUC-GAAACUAGUUGACAUC-----G--------CCGUGCCGC ....(((((((......(((..(((......))).)))...((((((((.(((((...((((....))))...))-)))..))))))))...-----)--------)).)))).. ( -25.90) >DroAna_CAF1 15975 110 - 1 UCAAA-CAGACAAACAACUG-UCGACCUAAUUCGAUGCAAAGUCAAUUAAUUCGAGCAGAUUAAUUAGUUUAUGGGGAAACUAGUUGACAUUUCAUCCGC---CGCCACGGCCGC .....-...........(((-((((..(((((.(((.....))))))))..)))).))).(((((((((((......)))))))))))..........((---((...))))... ( -23.20) >DroPer_CAF1 17728 112 - 1 GCCAA-CAGGCAAACAACUG-UCGACCUAAUUCGAUGCAAAGUCAAUUAAUUCGGGCAGAUUAAUUAGUUUAUUC-GAAACUAGUUGACAUUCCGUCCACACCGGGCUCGGCCGC (((..-..)))........(-(((((((.....((((.((.((((((((.((((((.(((((....))))).)))-)))..)))))))).)).))))......))).)))))... ( -29.80) >consensus GCAAA_CAGGCAAACAACUG_ACGACCUAAUUCGAUGCAAAGUCAAUUAAUUCGGGCAGAUUAAUUAGUUUAUUC_GAAACUAGUUGACAUU_C_UCC________CUCGGCCGC ........(((......(((.((((..(((((.(((.....))))))))..)))).))).(((((((((((......)))))))))))......................))).. (-19.08 = -19.97 + 0.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:26 2006