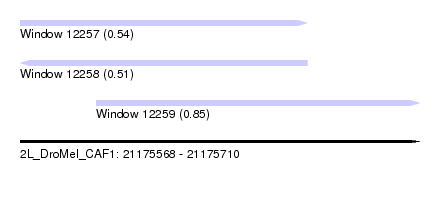
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,175,568 – 21,175,710 |
| Length | 142 |
| Max. P | 0.848285 |

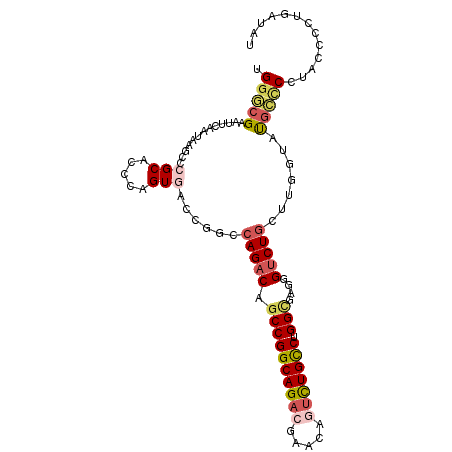
| Location | 21,175,568 – 21,175,670 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 83.73 |
| Mean single sequence MFE | -37.93 |
| Consensus MFE | -24.66 |
| Energy contribution | -25.58 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544132 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
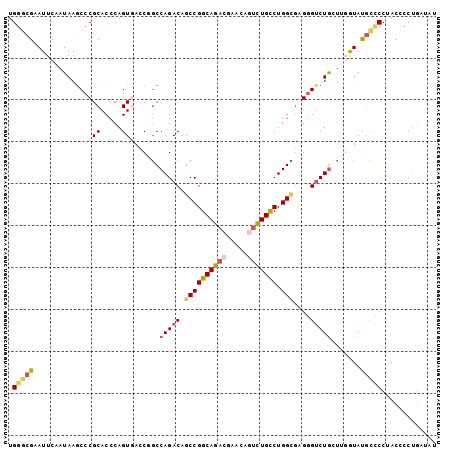
>2L_DroMel_CAF1 21175568 102 + 22407834 UGGGCGAAUUCUAUAAGCCCGCACCCAGUGACCGGCCAGACAGCCGGCAGACGAACAGUCUGCCUGGCGUGAGUCUGCUUGGUAUGCUCCUACCCCUGAUAU .((((...........))))......(((((((((.(((((.((((((((((.....))))))).)))....))))).))))).)))).............. ( -39.30) >DroSec_CAF1 12845 102 + 1 UGGGCGAAUUCUAUAAGCCCGCACCCAGUUACCGGCCAGACAGCCGGCAGACGAACAGUCUGCCUGGCGUGGGUCUGCUUUAUAUGCCCCUACCCCUGAUAU .(((((.((....((..(((((.((........(((......)))(((((((.....))))))).)).)))))..))....)).)))))............. ( -39.10) >DroSim_CAF1 13122 102 + 1 UGGGCGAAUUCUAUAAGCCCGCACCCAGUUACCGGCCAGACAGCCGGCAGACGAACAGUCUGCCUGGCGUGGGUCUGCUUUAUAUGCCCCUACCCCUGAUAU .(((((.((....((..(((((.((........(((......)))(((((((.....))))))).)).)))))..))....)).)))))............. ( -39.10) >DroEre_CAF1 14396 100 + 1 UGGACGAAUUCAAUAAGCCCGCACCCAGUGACC-GCCAGACAGCCGGCAGACGAACAGUCUGCCUGGUGAGGG-CUGGUUGGUAUGCCCCUACUCCUGAAGU .((.((...(((((.(((((.((((........-((......)).(((((((.....))))))).)))).)))-)).)))))..)))).............. ( -36.00) >DroYak_CAF1 14250 102 + 1 UGGGCGAAUUCAAUAAACCCGCACCCAGUGACCAGCCAGACAGCCGGCAGACGAACAAUCUGCCUGGUGAGGGUCUGGUUGGUAUGCCCCUACCCCUGAAAU .(((((.............(((.....)))(((((((((((.(((((((((.......)))))).)))....))))))))))).)))))............. ( -42.90) >DroAna_CAF1 12565 93 + 1 UGCCAGGAUUCAAUAAGCCCGCACCCAGUGAACUCCCAGACAGCCGGCAGACAA-----UUGUCUGGGCAGGGUCUAGUCGGUACAGUCCGACUCC----AU .....(((.....((..(((.(((...)))....(((((((((..(.....)..-----)))))))))..)))..))(((((......))))))))----.. ( -31.20) >consensus UGGGCGAAUUCAAUAAGCCCGCACCCAGUGACCGGCCAGACAGCCGGCAGACGAACAGUCUGCCUGGCGAGGGUCUGCUUGGUAUGCCCCUACCCCUGAUAU .(((((.............(((.....)))......(((((.((((((((((.....))))))).)))....))))).......)))))............. (-24.66 = -25.58 + 0.92)

| Location | 21,175,568 – 21,175,670 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 83.73 |
| Mean single sequence MFE | -40.08 |
| Consensus MFE | -27.62 |
| Energy contribution | -28.59 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.511949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21175568 102 - 22407834 AUAUCAGGGGUAGGAGCAUACCAAGCAGACUCACGCCAGGCAGACUGUUCGUCUGCCGGCUGUCUGGCCGGUCACUGGGUGCGGGCUUAUAGAAUUCGCCCA .......((((.(((...((..((((...(.(((.((((((((((.....))))))(((((....)))))....))))))).).)))).))...))))))). ( -39.20) >DroSec_CAF1 12845 102 - 1 AUAUCAGGGGUAGGGGCAUAUAAAGCAGACCCACGCCAGGCAGACUGUUCGUCUGCCGGCUGUCUGGCCGGUAACUGGGUGCGGGCUUAUAGAAUUCGCCCA .......((((..(..(.(((((.((.((((((.(((.(((((((.....)))))))((((....)))))))...))))).)..))))))))..)..)))). ( -43.20) >DroSim_CAF1 13122 102 - 1 AUAUCAGGGGUAGGGGCAUAUAAAGCAGACCCACGCCAGGCAGACUGUUCGUCUGCCGGCUGUCUGGCCGGUAACUGGGUGCGGGCUUAUAGAAUUCGCCCA .......((((..(..(.(((((.((.((((((.(((.(((((((.....)))))))((((....)))))))...))))).)..))))))))..)..)))). ( -43.20) >DroEre_CAF1 14396 100 - 1 ACUUCAGGAGUAGGGGCAUACCAACCAG-CCCUCACCAGGCAGACUGUUCGUCUGCCGGCUGUCUGGC-GGUCACUGGGUGCGGGCUUAUUGAAUUCGUCCA .............((((....(((..((-(((.((((.(((((((.....)))))))(((((.....)-))))....)))).)))))..))).....)))). ( -42.70) >DroYak_CAF1 14250 102 - 1 AUUUCAGGGGUAGGGGCAUACCAACCAGACCCUCACCAGGCAGAUUGUUCGUCUGCCGGCUGUCUGGCUGGUCACUGGGUGCGGGUUUAUUGAAUUCGCCCA ....(((.((((......)))).(((((.((..((((.(((((((.....))))))))).))...)))))))..)))((.((((((((...)))))))))). ( -39.20) >DroAna_CAF1 12565 93 - 1 AU----GGAGUCGGACUGUACCGACUAGACCCUGCCCAGACAA-----UUGUCUGCCGGCUGUCUGGGAGUUCACUGGGUGCGGGCUUAUUGAAUCCUGGCA ..----(((.((((.(((((((..((((((...((((((((..-----..)))))..))).)))))).((....)).))))))).....)))).)))..... ( -33.00) >consensus AUAUCAGGGGUAGGGGCAUACCAACCAGACCCACGCCAGGCAGACUGUUCGUCUGCCGGCUGUCUGGCCGGUCACUGGGUGCGGGCUUAUAGAAUUCGCCCA .............((((.........((.(((.((((.(((((((.....)))))))((((....))))........)))).)))))..........)))). (-27.62 = -28.59 + 0.97)

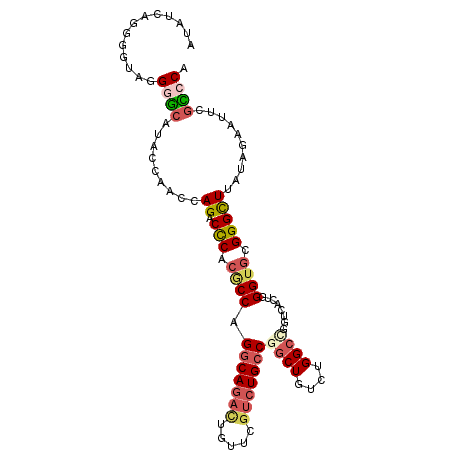
| Location | 21,175,595 – 21,175,710 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 73.85 |
| Mean single sequence MFE | -33.16 |
| Consensus MFE | -20.04 |
| Energy contribution | -20.60 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848285 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21175595 115 + 22407834 GUGACCGGCCAGACAGCCGGCAGACGAACAGUCUGCCUGGCGUGAGUCUGCUUGGUAUGCUCCUACCCCUGAUAUCUUCCUCUCAUUUCGUAGUCCCGCUCCUCUUAACCCCUAA ((((((((.(((((.((((((((((.....))))))).)))....))))).)))))......((((...(((..........)))....))))...)))................ ( -34.20) >DroSec_CAF1 12872 110 + 1 GUUACCGGCCAGACAGCCGGCAGACGAACAGUCUGCCUGGCGUGGGUCUGCUUUAUAUGCCCCUACCCCUGAUAUCCUCCUCUCCUUUCGUAGUCCCCCUCCUCUUAAGU----- ......((...(((.((((((((((.....))))))).)))(((((...((.......)).)))))..........................)))..))...........----- ( -30.20) >DroSim_CAF1 13149 110 + 1 GUUACCGGCCAGACAGCCGGCAGACGAACAGUCUGCCUGGCGUGGGUCUGCUUUAUAUGCCCCUACCCCUGAUAUCCUCCUCUCCUUUCGUAGUCCCCCUCCUCUUAAGU----- ......((...(((.((((((((((.....))))))).)))(((((...((.......)).)))))..........................)))..))...........----- ( -30.20) >DroEre_CAF1 14423 108 + 1 GUGACC-GCCAGACAGCCGGCAGACGAACAGUCUGCCUGGUGAGGG-CUGGUUGGUAUGCCCCUACUCCUGAAGUGUUCUUCCCGUGGUUUAGCCCCACUCCUCCCAAAU----- ((((((-(((((.(.((((((((((.....))))))).)))...).-)))).))))......(((..((.((((....))))....))..)))...)))...........----- ( -36.20) >DroYak_CAF1 14277 110 + 1 GUGACCAGCCAGACAGCCGGCAGACGAACAAUCUGCCUGGUGAGGGUCUGGUUGGUAUGCCCCUACCCCUGAAAUUUUCUUUCCCUGCUUUAGUCCCACUCCUCCCAAAU----- ((((((((((((((.(((((((((.......)))))).)))....)))))))))))............(((((...............)))))...)))...........----- ( -35.56) >DroAna_CAF1 12592 94 + 1 GUGAACUCCCAGACAGCCGGCAGACAA-----UUGUCUGGGCAGGGUCUAGUCGGUACAGUCCGACUCC----AUCCCCCGC--C-----AGGUCCUGUACCACUAAAAA----- (((.....((((((((..(.....)..-----))))))))(((((..((((((((......))))))..----.........--.-----))..)))))..)))......----- ( -32.60) >consensus GUGACCGGCCAGACAGCCGGCAGACGAACAGUCUGCCUGGCGAGGGUCUGCUUGGUAUGCCCCUACCCCUGAUAUCCUCCUCUCCUUUCGUAGUCCCACUCCUCUUAAAU_____ .........(((((.((((((((((.....))))))).)))....)))))................................................................. (-20.04 = -20.60 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:25 2006