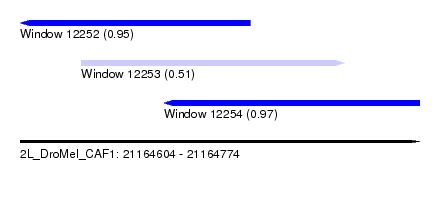
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,164,604 – 21,164,774 |
| Length | 170 |
| Max. P | 0.972090 |

| Location | 21,164,604 – 21,164,702 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 76.47 |
| Mean single sequence MFE | -16.63 |
| Consensus MFE | -12.97 |
| Energy contribution | -12.92 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21164604 98 - 22407834 ACAAU-UUUAAAGCCAAGAACAGUUUAGUUUAGAGUAGUUUAUAAACUGAUCUACACGUGGCCUAACAUAU---UAUCUAUCAUUACCAUUAAUUAUUCUAA-- .....-......((((.....((.((((((((..........)))))))).)).....)))).........---............................-- ( -12.40) >DroSim_CAF1 2116 101 - 1 UCAAU-UUUAAAGCCAAGAACAAUUUAGUUUAGAGCAGUUUAUAAACUGAUCUACACAUGGCUUUACAUAUUAUUAUCUAUCAUUACCAUUAAUUAUUCGAA-- .....-..((((((((.((((......))))(((.(((((....))))).))).....))))))))....................................-- ( -17.00) >DroEre_CAF1 3148 95 - 1 GCAAU-UUUAAAGCCAAGAACAGUUUAGUUUAGAGCAGUUUAUAAACUGAUCUACACGUGGCAUAACAUAUU------CAAUUUUACACCUAAUUAUUACAA-- .....-......((((...........((.((((.(((((....))))).)))).)).))))..........------........................-- ( -13.70) >DroYak_CAF1 3131 98 - 1 ACAAU-UUUAAAGCCAAGAACAGUUUAGUUUAGAGCAGUUUAUAAACUGAUCUACACGUGGCAUAACAUAUU---AUUUAUAAUUACACUUUAUUAUACUAA-- .....-......((((...........((.((((.(((((....))))).)))).)).))))..........---...((((((........))))))....-- ( -14.80) >DroMoj_CAF1 3552 96 - 1 GCAAA-GUUAAAGCCAAGAACAGUUCAGUUUAGCGUAGUUUAUAAACUGAUCUACACGUGGCUUGACAUUCA---AUUAAUUGUUUUUGA-AAU---GCAUUCU (((..-(((((.((((.....((.((((((((..........)))))))).)).....))))))))).((((---(..........))))-).)---))..... ( -23.90) >DroPer_CAF1 2143 91 - 1 AUAAUCUUUUAAGCCAAGAACAGUUCAGUUUAGAGCAGUCUAUAAACUGAUCUACACGUGGCCUAACA-----------AUAACUACCAUUUACUGUAGAUU-- ........(((.((((.....((.((((((((.((....)).)))))))).)).....)))).)))..-----------....((((........))))...-- ( -18.00) >consensus ACAAU_UUUAAAGCCAAGAACAGUUUAGUUUAGAGCAGUUUAUAAACUGAUCUACACGUGGCAUAACAUAUU___AU_UAUAAUUACCAUUAAUUAUUCAAA__ ............((((.....((.((((((((..........)))))))).)).....)))).......................................... (-12.97 = -12.92 + -0.05)


| Location | 21,164,630 – 21,164,742 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 81.51 |
| Mean single sequence MFE | -25.19 |
| Consensus MFE | -17.44 |
| Energy contribution | -17.25 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21164630 112 + 22407834 UA-AUAUGUUAGGCCACGUGUAGAUCAGUUUAUAAACUACUCUAAACUAAACUGUUCUUGGCUUUAAA-AUUGUUCUGGCAGCAUUGUAUCAGCGCUUAGGUAAACGCAUUUAA ..-..((((.((((((.....(((.(((((((...............))))))).)))))))))....-.....((((((.((.........)))).)))).....)))).... ( -17.16) >DroVir_CAF1 3508 108 + 1 U-AAAAUGUGAGGCCACGUGUAGAUCAGUUUAUAGACUACGCUAAACUGAACUGUUCCUGGCUUC-GU-UUUGCUCUGGCAGCAUUGUAUCAGCGCUCAAAUGAGCUUG---CA .-..((((((((((((.(..(((.((((((((..........)))))))).)))..).)))))))-..-..(((....))))))))......((((((....))))..)---). ( -33.50) >DroPse_CAF1 2115 107 + 1 ------UGUUAGGCCACGUGUAGAUCAGUUUAUAGACUGUUCUAAACUGAACUGUUCUUGGCUUAAAAGAUUAUUCUGGCAGCAUUGUAUCAGCAUUCAAUUAAGCUUAAU-CG ------..((((((((.(..(((.((((((..((((....)))))))))).)))..).))))))))..(((((..((........(((....)))........))..))))-). ( -26.69) >DroEre_CAF1 3172 111 + 1 --AAUAUGUUAUGCCACGUGUAGAUCAGUUUAUAAACUGCUCUAAACUAAACUGUUCUUGGCUUUAAA-AUUGCUAUUGCAGCAUUGUAUCAGCGCUUAGAUAAACUCAUUUCA --.((((((......))))))((((.(((((((.....((.((..((....((((...((((......-...))))..))))....))...)).))....))))))).)))).. ( -19.30) >DroMoj_CAF1 3576 109 + 1 U-UGAAUGUCAAGCCACGUGUAGAUCAGUUUAUAAACUACGCUAAACUGAACUGUUCUUGGCUUUAAC-UUUGCUCUGGCAGCAUUGUAUCAGCGCUCAAAUAAACUGA---CA .-....((((((((((.(..(((.((((((((..........)))))))).)))..).))))))..((-..((((.....))))..))...................))---)) ( -26.10) >DroPer_CAF1 2166 107 + 1 ------UGUUAGGCCACGUGUAGAUCAGUUUAUAGACUGCUCUAAACUGAACUGUUCUUGGCUUAAAAGAUUAUUCUGGCAGCAUUGUAUCAGCAUUCAAUUAAGCUUAAU-CG ------..((((((((.(..(((.((((((((.((....)).)))))))).)))..).))))))))..(((((..((........(((....)))........))..))))-). ( -28.39) >consensus ___A_AUGUUAGGCCACGUGUAGAUCAGUUUAUAAACUACUCUAAACUGAACUGUUCUUGGCUUUAAA_AUUGCUCUGGCAGCAUUGUAUCAGCGCUCAAAUAAACUUA_U_CA ..........((((((.(..(((.((((((((..........)))))))).)))..).))))))........((.(((((......))..))).)).................. (-17.44 = -17.25 + -0.19)

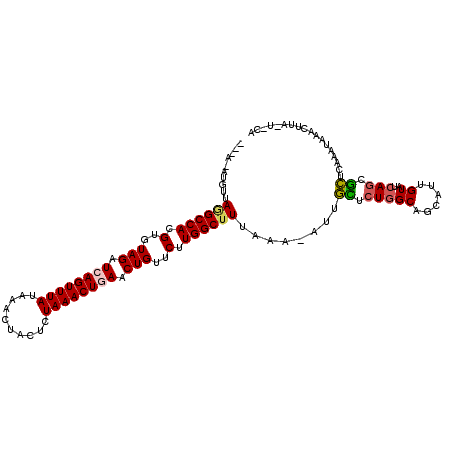
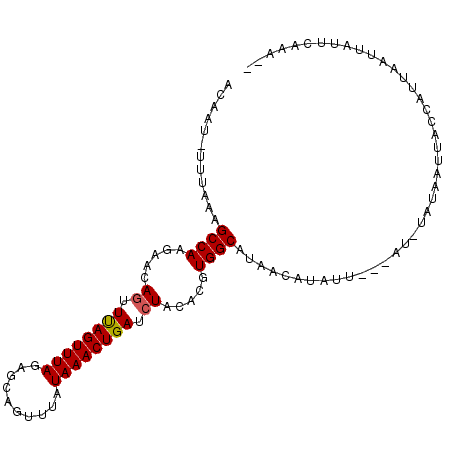
| Location | 21,164,665 – 21,164,774 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 76.82 |
| Mean single sequence MFE | -26.01 |
| Consensus MFE | -21.60 |
| Energy contribution | -20.74 |
| Covariance contribution | -0.86 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21164665 109 - 22407834 CA----AAACA--AGCAGCAACAAGAAAGUGCUUAGGCUUAAAUGCGUUUACCUAAGCGCUGAUACAAUGCUGCCAGAACAAU-UUUAAAGCCAAGAACAGUUUAGUUUAGAGUAG ..----((((.--.((((((.......((((((((((..((((....)))))))))))))).......))))))..((((..(-(((.......))))..)))).))))....... ( -27.14) >DroVir_CAF1 3543 101 - 1 --------UCC--AGCAGCAAAAAGAAAGCGUUCAAAUUG---CAAGCUCAUUUGAGCGCUGAUACAAUGCUGCCAGAGCAAA-AC-GAAGCCAGGAACAGUUCAGUUUAGCGUAG --------((.--.((((((.......(((((((((((.(---....)..))))))))))).......))))))..))((.((-((-..(((........)))..)))).)).... ( -30.34) >DroPse_CAF1 2145 111 - 1 CA----AUCCGAUGGCAGCAAAAAGAAAGUGUUCAAAUCG-AUUAAGCUUAAUUGAAUGCUGAUACAAUGCUGCCAGAAUAAUCUUUUAAGCCAAGAACAGUUCAGUUUAGAACAG ..----......((((((((.......(((((((((....-...........))))))))).......))))))))......((((.......))))...((((......)))).. ( -25.00) >DroYak_CAF1 3192 109 - 1 CA----UAAUA--AGCAGCAACAAGAAAGUGCUUAAGUUGAAAUGGGUUUAUCUAAGCGCUGAUACAAUGCUGCCAGAACAAU-UUUAAAGCCAAGAACAGUUUAGUUUAGAGCAG ..----.....--.((((((.......((((((((.(.((((.....)))).))))))))).......)))))).........-......((...((((......))))...)).. ( -22.94) >DroMoj_CAF1 3611 110 - 1 UGAUGCUAAUC--AGCAGCAACAAGAAAGUCUUCAAAUUG---UCAGUUUAUUUGAGCGCUGAUACAAUGCUGCCAGAGCAAA-GUUAAAGCCAAGAACAGUUCAGUUUAGCGUAG ..(((((((.(--.((((((.......(((.(((((((..---.......))))))).))).......))))))..((((...-(((.........))).)))).).))))))).. ( -25.64) >DroPer_CAF1 2196 111 - 1 CA----AUCCGAUGGCAGCAAAAAGAAAGUGUUCAAAUCG-AUUAAGCUUAAUUGAAUGCUGAUACAAUGCUGCCAGAAUAAUCUUUUAAGCCAAGAACAGUUCAGUUUAGAGCAG ..----......((((((((.......(((((((((....-...........))))))))).......))))))))......((((.......))))...((((......)))).. ( -25.00) >consensus CA____AAACA__AGCAGCAAAAAGAAAGUGUUCAAAUUG_A_UAAGCUUAUUUGAGCGCUGAUACAAUGCUGCCAGAACAAU_UUUAAAGCCAAGAACAGUUCAGUUUAGAGCAG ..............((((((.......(((((((((((............))))))))))).......))))))................((...((((......))))...)).. (-21.60 = -20.74 + -0.86)

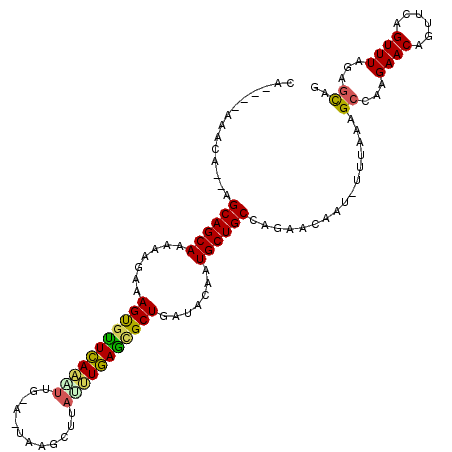
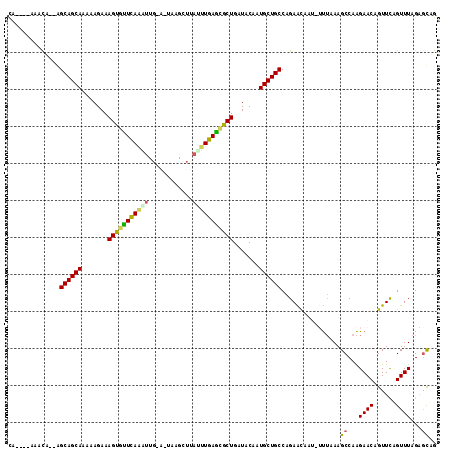
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:21 2006