
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,150,610 – 21,150,723 |
| Length | 113 |
| Max. P | 0.999985 |

| Location | 21,150,610 – 21,150,703 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 71.90 |
| Mean single sequence MFE | -21.65 |
| Consensus MFE | -9.31 |
| Energy contribution | -10.88 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.632272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21150610 93 + 22407834 CGUGCCACUGCAGAGCGAAGCAAGUGCAAUUGCCAAUUGCAAAUGCAAUUGACGGUAAAGGCGAACAAACUUCGAUGGAAAUAUCGAUUUCAA--- ..((((..(((...((((.((....))..))))(((((((....)))))))...)))..))))........((((((....))))))......--- ( -23.80) >DroSec_CAF1 8545 93 + 1 CGUGUCACUGCAAAUCGAAGCAAGUGCAAUUGCAAAUUACAAACGCAAUUGAGGGUAAAGGUGAAGAAACAUCGAUGGAAAUAUCGAUUUCAA--- ....(((((((........)).....(((((((...........)))))))........))))).(((..(((((((....))))))))))..--- ( -20.00) >DroEre_CAF1 8899 77 + 1 CGUGCGACUGCUAAGCGAAGCAAGUGCAAUUGCCAAU-------------------GGAGGUGAUGAAACAUCGAAGGAAACAUCGAUUUGGAAAA ..((((..((((......))))..))))((..((...-------------------...))..)).....(((((.(....).)))))........ ( -20.50) >DroYak_CAF1 8569 89 + 1 -------CUACUAAGCGAAGCAAGUGCAAUUGGCAAUUGCAAAUGCAAUUGACUGUAGAGGUGAUAAAACAUCGAUGGAACCAUCGAUUUUAAAAA -------((((....(((.(((..((((((.....))))))..)))..)))...))))............(((((((....)))))))........ ( -22.30) >consensus CGUGCCACUGCAAAGCGAAGCAAGUGCAAUUGCCAAUUGCAAAUGCAAUUGACGGUAAAGGUGAAGAAACAUCGAUGGAAACAUCGAUUUCAA___ ........(((........((((......))))(((((((....))))))).........)))........((((((....))))))......... ( -9.31 = -10.88 + 1.56)

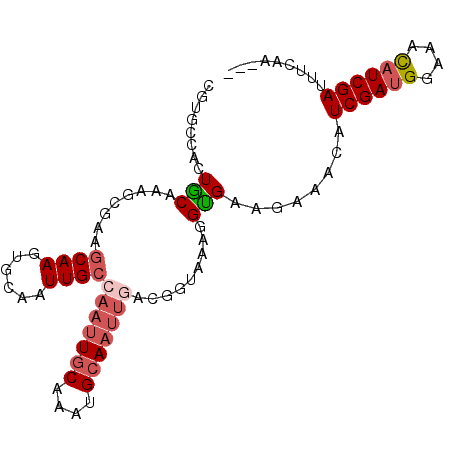
| Location | 21,150,632 – 21,150,723 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 72.73 |
| Mean single sequence MFE | -27.03 |
| Consensus MFE | -15.47 |
| Energy contribution | -16.48 |
| Covariance contribution | 1.01 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.47 |
| Structure conservation index | 0.57 |
| SVM decision value | 4.81 |
| SVM RNA-class probability | 0.999952 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21150632 91 + 22407834 AGUGCAAUUGCCAAUUGCAAAUGCAAUUGACGGUAAAGGCGAACAAACUUCGAUGGAAAUAUCGAUUUCAA-----------AAACAUCGAUGUUUCAGUGA------------ ..(((..(((((((((((....))))))...)))))..)))........(((...(((((((((((.....-----------....)))))))))))..)))------------ ( -25.30) >DroSec_CAF1 8567 91 + 1 AGUGCAAUUGCAAAUUACAAACGCAAUUGAGGGUAAAGGUGAAGAAACAUCGAUGGAAAUAUCGAUUUCAA-----------AAACAUCGAUGUUUCAGCGA------------ .((.(((((((...........)))))))..............(((((((((((((((((....)))))..-----------...)))))))))))).))..------------ ( -25.20) >DroYak_CAF1 8584 114 + 1 AGUGCAAUUGGCAAUUGCAAAUGCAAUUGACUGUAGAGGUGAUAAAACAUCGAUGGAACCAUCGAUUUUAAAAAUUAUCGAAAAUCAUCGAAGUUUUAGUAUUGGCCUGCGACU (((((....(((((((((....))))))).))....((((.(((((((.(((((((.....(((((..........)))))...))))))).))))))....).)))))).))) ( -30.60) >consensus AGUGCAAUUGCCAAUUGCAAAUGCAAUUGACGGUAAAGGUGAAAAAACAUCGAUGGAAAUAUCGAUUUCAA___________AAACAUCGAUGUUUCAGUGA____________ ..(((.((((.(((((((....))))))).)))).........(((((.((((((..............................)))))).))))).)))............. (-15.47 = -16.48 + 1.01)

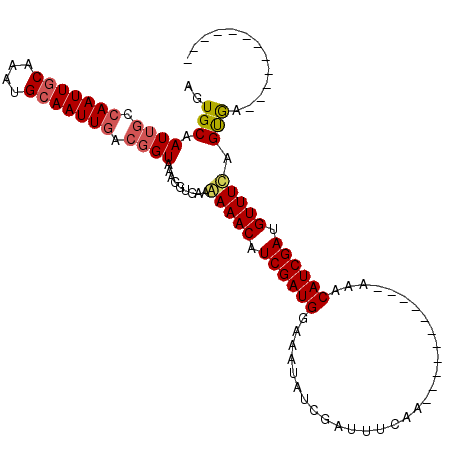

| Location | 21,150,632 – 21,150,723 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 72.73 |
| Mean single sequence MFE | -24.90 |
| Consensus MFE | -14.92 |
| Energy contribution | -15.81 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.59 |
| Structure conservation index | 0.60 |
| SVM decision value | 5.38 |
| SVM RNA-class probability | 0.999985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21150632 91 - 22407834 ------------UCACUGAAACAUCGAUGUUU-----------UUGAAAUCGAUAUUUCCAUCGAAGUUUGUUCGCCUUUACCGUCAAUUGCAUUUGCAAUUGGCAAUUGCACU ------------......((((.((((((...-----------..(((((....))))))))))).)))).............(((((((((....)))))))))......... ( -22.30) >DroSec_CAF1 8567 91 - 1 ------------UCGCUGAAACAUCGAUGUUU-----------UUGAAAUCGAUAUUUCCAUCGAUGUUUCUUCACCUUUACCCUCAAUUGCGUUUGUAAUUUGCAAUUGCACU ------------..(..((((((((((((...-----------..(((((....)))))))))))))))))..)...........((((((((.........)))))))).... ( -24.50) >DroYak_CAF1 8584 114 - 1 AGUCGCAGGCCAAUACUAAAACUUCGAUGAUUUUCGAUAAUUUUUAAAAUCGAUGGUUCCAUCGAUGUUUUAUCACCUCUACAGUCAAUUGCAUUUGCAAUUGCCAAUUGCACU (((.((((........((((((.((((......))))...........(((((((....)))))))))))))...........(.(((((((....))))))).)..))))))) ( -27.90) >consensus ____________UCACUGAAACAUCGAUGUUU___________UUGAAAUCGAUAUUUCCAUCGAUGUUUCUUCACCUUUACCGUCAAUUGCAUUUGCAAUUGGCAAUUGCACU .................(((((.((((((..............................)))))).)))))............(((((((((....)))))))))......... (-14.92 = -15.81 + 0.89)

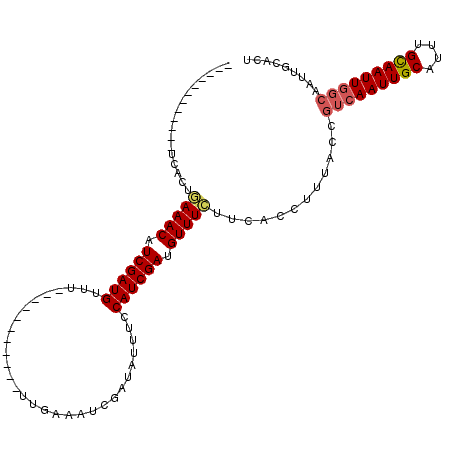
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:18 2006