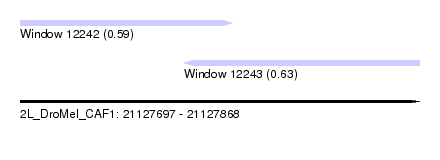
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,127,697 – 21,127,868 |
| Length | 171 |
| Max. P | 0.634938 |

| Location | 21,127,697 – 21,127,788 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 78.83 |
| Mean single sequence MFE | -20.58 |
| Consensus MFE | -14.34 |
| Energy contribution | -14.40 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.590253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
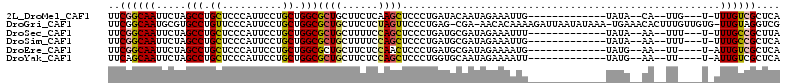
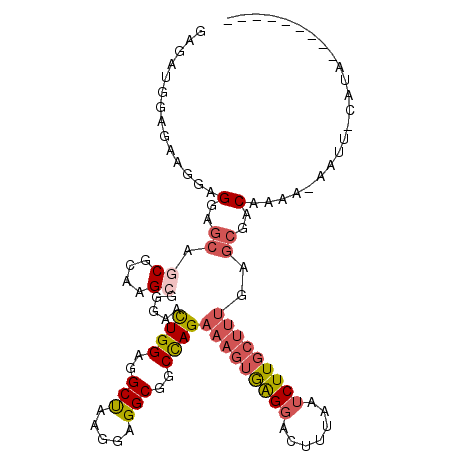
>2L_DroMel_CAF1 21127697 91 + 22407834 UUCGGCAAUUCUAGCCUGCUCCCAUUCCUGCUGGCGCUGCUUCUCAAGCUCCCUGAUACAAUAGAAAUUG-------------UAUA--CA--UUG---U-UUUGUCGCUCA ..((((((..(..(((.((..........)).)))(((........)))....(((((((((....))))-------------))).--))--..)---.-.)))))).... ( -20.50) >DroGri_CAF1 3345 108 + 1 UUCGGCAAUGCGUGCCUGUUCCCAUUCCUGCUGGCGCUGCUUCUCUAGUUCCCUGAG-CGA-AACACAAAAGAUUAAUAUAAA-UGAAACACUUUGUUGUG-UUGUAGGUCG ..((((...(((((((.((..........)).))))).))....(((((((...)))-)..-.....................-...(((((......)))-)).))))))) ( -20.70) >DroSec_CAF1 2601 91 + 1 UUCGGCAAUUCUAGCCUGCUCCCAUUCCUGCUGGCGCUGCUUUUCCAGCUCCCUGAUGCGAUAGAAAUUU-------------UAUA--AA--UUU---U-UUUGCCGCUUA ..((((((((((((((.((..........)).)))((((......))))............)))))....-------------....--..--...---.-.)))))).... ( -20.46) >DroSim_CAF1 2599 91 + 1 UUCGGCAAUUCUAGCCUGCUCCCAUUCCUGCUGGCGCUGCUUUUCCAGCUCCCUGAUGCGAUAGAAAUUG-------------UAUA--AA--UUU---U-UUUGCCGCUCA ..((((((.....(((.((..........)).)))((((......))))......(((((((....))))-------------))).--..--...---.-.)))))).... ( -23.30) >DroEre_CAF1 2569 90 + 1 UUCGGCAAUUCUAGCCUGCUCCCAUUCCUGCUGGCGCUGCUUCUCCAACUCCCUGAUGCGAUAGAAAAUG-------------UAUG--AA--UU----U-AUUGUCGCUCA ...((((......(((.((..........)).)))..)))).............((.(((((((.((((.-------------....--.)--))----)-.))))))))). ( -19.60) >DroYak_CAF1 2547 90 + 1 UUCAGCAAUUCUAGCCUGCUCCCAUUCCUGCUGGCGCUGCUUCUCCAGCUCCCUGGUGCAAUAGAAAAUU-------------UAUG--AA--UU----U-AUUGUCGCUCA ..(((........(((.((..........)).)))((((......))))...)))((((((((((.....-------------....--..--))----)-)))).)))... ( -18.90) >consensus UUCGGCAAUUCUAGCCUGCUCCCAUUCCUGCUGGCGCUGCUUCUCCAGCUCCCUGAUGCGAUAGAAAAUG_____________UAUA__AA__UU____U_UUUGUCGCUCA ..((((((.....(((.((..........)).)))((((......)))).....................................................)))))).... (-14.34 = -14.40 + 0.06)
| Location | 21,127,767 – 21,127,868 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 83.06 |
| Mean single sequence MFE | -24.33 |
| Consensus MFE | -17.11 |
| Energy contribution | -18.00 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
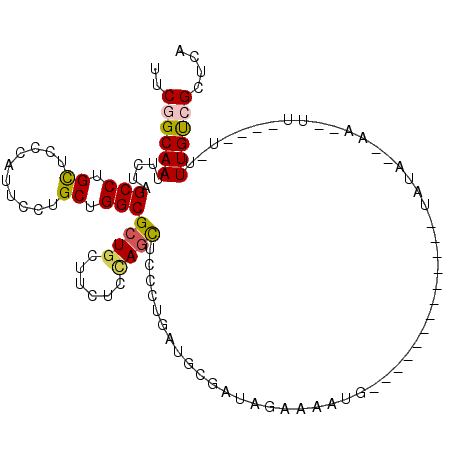
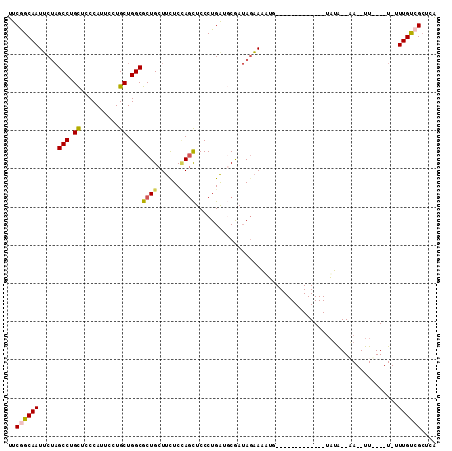
>2L_DroMel_CAF1 21127767 101 - 22407834 GAGAUGGAGAAGGAGGAGCAGCGCAAGAGGGAACUGGAGGCUAAGGAGGCGGCCAGAAAGUAAGGACUUUUAUCUUGCUUUGAGCGACAAAACAAUG-UAUA--------- ...............(..(((.((((((.....((((..(((.....)))..))))(((((....)))))..)))))).)))..)............-....--------- ( -22.30) >DroSec_CAF1 2671 101 - 1 GAGAUGGAGAUGGAGGAGCAGCGCAAGCGGGAACUGGAGGCUAAGGAGGCGGCCAGAAAGUGAGGACUUUCAUCUUGCUUUAAGCGGCAAAAAAAUU-UAUA--------- ......((((((((((....((....)).....((((..(((.....)))..))))..........))))))))))(((......))).........-....--------- ( -28.60) >DroSim_CAF1 2669 101 - 1 GAGAUGGAAAUGGAGGAGCAGCGCAAGCGGGAACUGGAGGCUAAGGAGGCGGCCAGAAAGUGAGGAUUUUUAUCUUGCUUUGAGCGGCAAAAAAAUU-UAUA--------- (((((((((((......((.((....)).....((((..(((.....)))..))))...))....)))))))))))(((......))).........-....--------- ( -25.30) >DroEre_CAF1 2639 100 - 1 GAGAUGGAGAAGGAGGAGCAGCGCAAGCGGGAACUGGAGGCUAAGGAGGCAGCUAGAAAGUGAGGACAUUAAUCUUGCUUUGAGCGACAAUA-AAUU-CAUA--------- ...(((((......(..((.((....)).....((((..(((.....)))..))))((((..(((.......)))..))))..))..)....-..))-))).--------- ( -25.00) >DroYak_CAF1 2617 100 - 1 GAGAUGGAGAAGGAGGAGCAGCGCAAGCGGGAACUGGAGGCUAAGGAGGCGGCCAGAAAGUGAGGACAUUAAUCUUGCUUUGAGCGACAAUA-AAUU-CAUA--------- ...(((((......(..((.((....)).....((((..(((.....)))..))))((((..(((.......)))..))))..))..)....-..))-))).--------- ( -26.80) >DroAna_CAF1 3611 96 - 1 GAAAUGGAGAAGGAGGAGCAGCGCAAGAGGGAAUUGGAAGCCAAGGAGGCGGCCAGAAAGUAGG----UUAAUCUU-----------CAAGACUAUGACAUACUUAUUAUA (..((((.....(((((.(..(....)..)...((((..(((.....)))..))))........----....))))-----------)....))))..)............ ( -18.00) >consensus GAGAUGGAGAAGGAGGAGCAGCGCAAGCGGGAACUGGAGGCUAAGGAGGCGGCCAGAAAGUGAGGACUUUAAUCUUGCUUUGAGCGACAAAA_AAUU_CAUA_________ ..............(..((.((....)).....((((..(((.....)))..))))(((((((((.......)))))))))..))..)....................... (-17.11 = -18.00 + 0.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:11 2006