| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,113,884 – 21,113,993 |
| Length | 109 |
| Max. P | 0.650184 |

| Location | 21,113,884 – 21,113,993 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 79.19 |
| Mean single sequence MFE | -35.60 |
| Consensus MFE | -22.34 |
| Energy contribution | -23.70 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.54 |
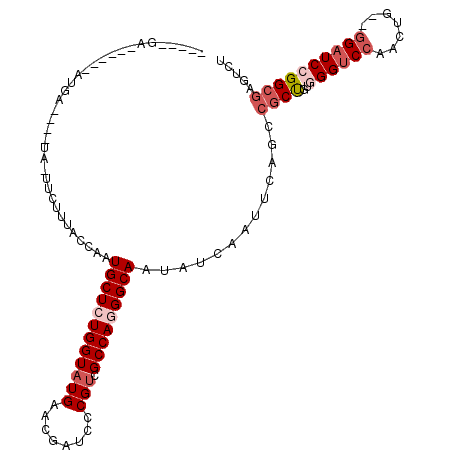
| Structure conservation index | 0.63 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21113884 109 - 22407834 GCUCUGAAU----AUGAUGGA--UGUCUUUACCAAUGCUCUGGUAUGAACGAUCCCGCCGCCACGGCAAUAUCAAUUCGGCGGCCGUGGGCCCAAUGGCCUGCUCCGGGGACUCU (((((((((----.(((((((--(((...(((((......)))))...)).)))).((((...))))...)))))))))).))).(..((((....))))..)...(....)... ( -37.60) >DroVir_CAF1 38015 96 - 1 --------------UGA----UA-UUGUUUACCAAUGCUCUGGUAUGAACGAUCCCGUCGCCAGGGCAGGAUCAGUUCGGCCGCUGUGGGUCCAACUGACGGAUCGGGCGAGUCU --------------.((----(.-((((((.((..((((((((((((........))).)))))))))...((((((.((((......)))).)))))).))...))))))))). ( -33.30) >DroPse_CAF1 14405 98 - 1 ----------UGGAUGA----UAAUCUUUUACCAAUGCUCUGGUAUGAACGAUCCCGUCGCCAGGGCAAUAUCAAUUCAGCCGCUGUGGGUCCAACU---GGAUCCGGCGAGUCU ----------(((((((----((............((((((((((((........))).))))))))).)))).)))))((((((..((((((....---)))))))))).)).. ( -31.82) >DroWil_CAF1 18059 103 - 1 --UUUUA------AUGG----UUUUGCUUUACCAAUGCUCUGGUAUGAACGAUCCCGUCGCCAUGGCAAAAUCAAUUCAGCCGCUGUGGGUCCAACUGAUGGAUCCGGCGAGUCU --.....------.(((----..(((...(((((......)))))....)))..)))(((((..(((............))).....(((((((.....)))))))))))).... ( -32.30) >DroMoj_CAF1 27138 105 - 1 -----GAUUCUUGGUGA----UA-UUGUUUACCAAUGCUCUGGUAUGAACGAUCCCGUCGCCAGGGCAGGAUCAGUUCGGCCGCUGUGGGUCCAACUGGUGGAUCGGGCGAGUCU -----((((((((((((----..-....))))))).((((.(((.((((((((((.(((.....))).))))).))))))))......((((((.....))))))))))))))). ( -42.80) >DroPer_CAF1 14334 106 - 1 --UCUGAAUCUGGAUGA----UAAUCUUUUACCAAUGCUCUGGUAUGAACGAUCCCGUCGCCAGGGCAAUAUCAAUUCAGCCGCUGUGGGUCCAACU---GGAUCCGGCGAGUCU --.((((((.(((.(((----.......)))))).((((((((((((........))).)))))))))......)))))).((((..((((((....---))))))))))..... ( -35.80) >consensus _____GA______AUGA____UA_UUCUUUACCAAUGCUCUGGUAUGAACGAUCCCGUCGCCAGGGCAAUAUCAAUUCAGCCGCUGUGGGUCCAACUG__GGAUCCGGCGAGUCU ...................................((((((((((((........))).))))))))).............((((..((((((.......))))))))))..... (-22.34 = -23.70 + 1.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:10 2006