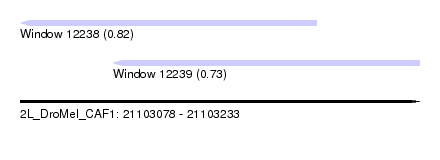
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,103,078 – 21,103,233 |
| Length | 155 |
| Max. P | 0.821961 |

| Location | 21,103,078 – 21,103,193 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 95.91 |
| Mean single sequence MFE | -26.53 |
| Consensus MFE | -25.49 |
| Energy contribution | -25.93 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21103078 115 - 22407834 GAAAUCAUGUGAUUGCUUCUUAGAUAACCAUUUUCAGAGGUUGUCACACAACUGGCAUGAUAUCAGGAAGUUGUCGCUUUUGAUGUACAUUUCUACAGCUCAUAAAUAAUACAAA ...((((.(((((.(((((...(((((((.........)))))))......((((.......))))))))).)))))...))))............................... ( -24.50) >DroSec_CAF1 10341 112 - 1 GAAAUCAUGUGAUUGCUUCUUAGAUAACCAUUUUCAGAGGUUGUCUGACAACUGACAUGAUAUCAGGAAGUUGUCGCUUUUGAUGUACCUUUCUACAGCUCAUAAAUAAUA---A ...((((.(((((.(((((((((((((((.........))))))))))...((((.......))))))))).)))))...))))...........................---. ( -28.00) >DroSim_CAF1 12850 112 - 1 GAAAUCAUGUGAUUGCUUCUUAGAUAACCAUUUUCAGAGGUUGUCUGACAACUGGCAUGAUAUCAGGAAGUUGUCGCUUUUGAUGUACAUUUCUACAGCUCAUAAAUAAUA---A ...((((.(((((.(((((((((((((((.........))))))))))...((((.......))))))))).)))))...))))...........................---. ( -27.10) >consensus GAAAUCAUGUGAUUGCUUCUUAGAUAACCAUUUUCAGAGGUUGUCUGACAACUGGCAUGAUAUCAGGAAGUUGUCGCUUUUGAUGUACAUUUCUACAGCUCAUAAAUAAUA___A ...((((.(((((.(((((((((((((((.........))))))))))...((((.......))))))))).)))))...))))............................... (-25.49 = -25.93 + 0.45)


| Location | 21,103,114 – 21,103,233 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 97.76 |
| Mean single sequence MFE | -27.70 |
| Consensus MFE | -26.59 |
| Energy contribution | -27.03 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725174 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21103114 119 - 22407834 GAUGGCUAUAUAAAUCUAUCUUUUAUUUAAACUAUGAUAAGAAAUCAUGUGAUUGCUUCUUAGAUAACCAUUUUCAGAGGUUGUCACACAACUGGCAUGAUAUCAGGAAGUUGUCGCUU (((((..........)))))(((((((........)))))))......(((((.(((((...(((((((.........)))))))......((((.......))))))))).))))).. ( -25.50) >DroSec_CAF1 10374 119 - 1 GAUGGCUAUAUAAAUCUAUCUUUUAUUUGAACUAUGAUAAGAAAUCAUGUGAUUGCUUCUUAGAUAACCAUUUUCAGAGGUUGUCUGACAACUGACAUGAUAUCAGGAAGUUGUCGCUU (((((..........)))))(((((((........)))))))......(((((.(((((((((((((((.........))))))))))...((((.......))))))))).))))).. ( -29.00) >DroSim_CAF1 12883 119 - 1 GAUGGCUAUAUAAAUCUAUCUUUUAUUUGAACUAUGAUAAGAAAUCAUGUGAUUGCUUCUUAGAUAACCAUUUUCAGAGGUUGUCUGACAACUGGCAUGAUAUCAGGAAGUUGUCGCUU ((..(((...............(((((........)))))((.(((((((.((((....((((((((((.........))))))))))))).).))))))).))....)))..)).... ( -28.60) >consensus GAUGGCUAUAUAAAUCUAUCUUUUAUUUGAACUAUGAUAAGAAAUCAUGUGAUUGCUUCUUAGAUAACCAUUUUCAGAGGUUGUCUGACAACUGGCAUGAUAUCAGGAAGUUGUCGCUU ((..(((...............(((((........)))))((.(((((((.((((....((((((((((.........))))))))))))).).))))))).))....)))..)).... (-26.59 = -27.03 + 0.45)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:08 2006