
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,092,266 – 21,092,357 |
| Length | 91 |
| Max. P | 0.973589 |

| Location | 21,092,266 – 21,092,357 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.58 |
| Mean single sequence MFE | -22.86 |
| Consensus MFE | -16.70 |
| Energy contribution | -17.22 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973589 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
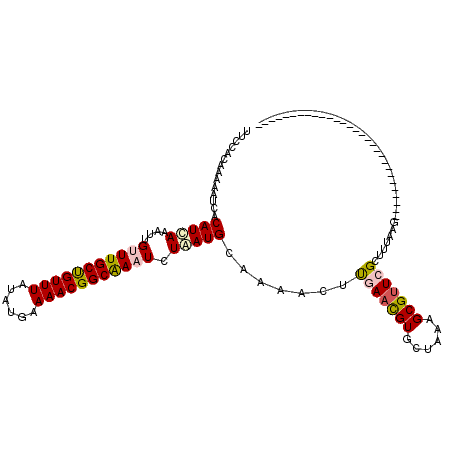
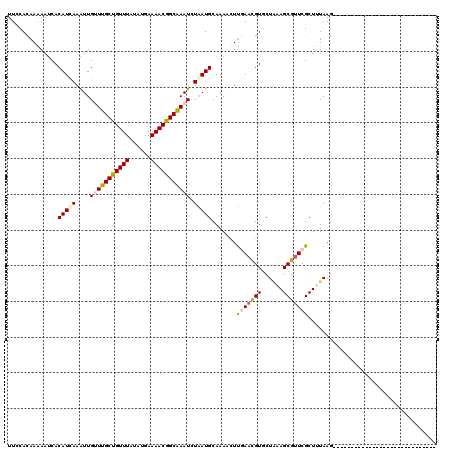
>2L_DroMel_CAF1 21092266 91 + 22407834 UUCCACAAGAAUCACAUCAAGUUGUUUGCCGUUUACAUGAAAACGGCGAAUUUGAUGCAAAAUUCGAACGUGCUAAAGCGUUCGCUGUAAG----------------------------- ....(((.((((..((((((...(((((((((((......)))))))))))))))))....))))((((((......))))))..)))...----------------------------- ( -28.60) >DroPse_CAF1 19246 91 + 1 UUCCACAAAAAUCACAUCAAAUUGGUUGCUGUUUAUAUGAAAACGGCAAAUCUAAUGCAAAUCUUGAAUGUGCUGAAGCGUUCGCUUUAAG----------------------------- ............((((((((((((((((((((((......)))))))))..))))).......))).))))).((((((....))))))..----------------------------- ( -20.31) >DroWil_CAF1 25972 120 + 1 UUUCAUAAAAAUCACAUUAAAUUGUUUGCUGUUUAUAUGAAAACGGCAAAUCUAAUGCAAAAUUUCAACGUGCUAAAGCGUUCGCUUUAAAAAGAGACGAGACGAGUGAGCGAGAGGCGA ((((..........(((((....(((((((((((......))))))))))).)))))..............(((....((((((((((....)))).))).)))....)))))))..... ( -26.30) >DroAna_CAF1 16506 91 + 1 UUCCAUAAGAACCACAUCAAGUUGUUUGCUGUUUAUAUGAAAACGGCAAAUCUGAUGCAAAACUUUAACGUCCUUAAGCGAUCGCUAUAAG----------------------------- ..............(((((....(((((((((((......))))))))))).)))))...............(((((((....))).))))----------------------------- ( -18.80) >DroPer_CAF1 19225 91 + 1 UUCCACAAAAAUCACAUCAAAUUGGUUGCUGUUUAUAUGAAAACGGCAAAUCUAAUGCAAAUCUUGAAUGUGCUGAAGCGUUCGCUUUAAG----------------------------- ............((((((((((((((((((((((......)))))))))..))))).......))).))))).((((((....))))))..----------------------------- ( -20.31) >consensus UUCCACAAAAAUCACAUCAAAUUGUUUGCUGUUUAUAUGAAAACGGCAAAUCUAAUGCAAAACUUGAACGUGCUAAAGCGUUCGCUUUAAG_____________________________ ..............(((((....(((((((((((......))))))))))).))))).......(((((((......))))))).................................... (-16.70 = -17.22 + 0.52)

| Location | 21,092,266 – 21,092,357 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.58 |
| Mean single sequence MFE | -22.82 |
| Consensus MFE | -16.44 |
| Energy contribution | -15.64 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961983 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
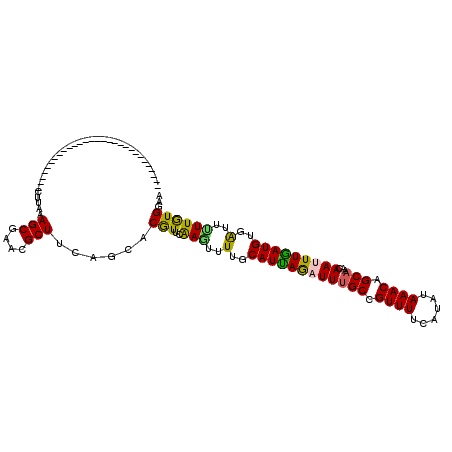
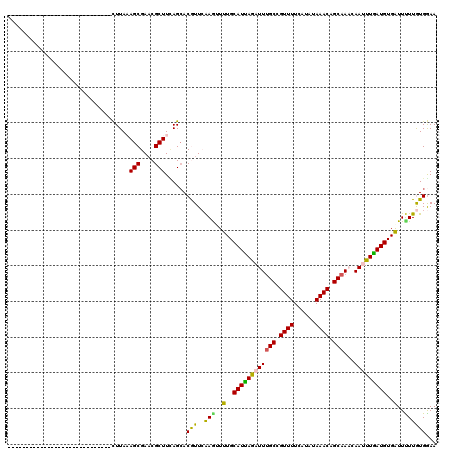
>2L_DroMel_CAF1 21092266 91 - 22407834 -----------------------------CUUACAGCGAACGCUUUAGCACGUUCGAAUUUUGCAUCAAAUUCGCCGUUUUCAUGUAAACGGCAAACAACUUGAUGUGAUUCUUGUGGAA -----------------------------.((((((.(((((........)))))(((((..(((((((.((.(((((((......)))))))....)).)))))))))))))))))).. ( -25.90) >DroPse_CAF1 19246 91 - 1 -----------------------------CUUAAAGCGAACGCUUCAGCACAUUCAAGAUUUGCAUUAGAUUUGCCGUUUUCAUAUAAACAGCAACCAAUUUGAUGUGAUUUUUGUGGAA -----------------------------....((((....)))).....(...(((((.(..((((((((((((.((((......)))).)))...)))))))))..).)))))..).. ( -21.10) >DroWil_CAF1 25972 120 - 1 UCGCCUCUCGCUCACUCGUCUCGUCUCUUUUUAAAGCGAACGCUUUAGCACGUUGAAAUUUUGCAUUAGAUUUGCCGUUUUCAUAUAAACAGCAAACAAUUUAAUGUGAUUUUUAUGAAA .........(((....(((.(((.((........))))))))....))).(((.((((((..(((((((((((((.((((......)))).)))...)))))))))))))))).)))... ( -23.80) >DroAna_CAF1 16506 91 - 1 -----------------------------CUUAUAGCGAUCGCUUAAGGACGUUAAAGUUUUGCAUCAGAUUUGCCGUUUUCAUAUAAACAGCAAACAACUUGAUGUGGUUCUUAUGGAA -----------------------------((((.(((....)))))))..(((..(((..(..((((((.(((((.((((......)))).)))))....))))))..)..))))))... ( -22.20) >DroPer_CAF1 19225 91 - 1 -----------------------------CUUAAAGCGAACGCUUCAGCACAUUCAAGAUUUGCAUUAGAUUUGCCGUUUUCAUAUAAACAGCAACCAAUUUGAUGUGAUUUUUGUGGAA -----------------------------....((((....)))).....(...(((((.(..((((((((((((.((((......)))).)))...)))))))))..).)))))..).. ( -21.10) >consensus _____________________________CUUAAAGCGAACGCUUCAGCACGUUCAAGUUUUGCAUUAGAUUUGCCGUUUUCAUAUAAACAGCAAACAAUUUGAUGUGAUUUUUGUGGAA ..................................(((....)))......(((..(((..(..((((((((((((.((((......)))).)))...)))))))))..)..))))))... (-16.44 = -15.64 + -0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:04 2006