
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
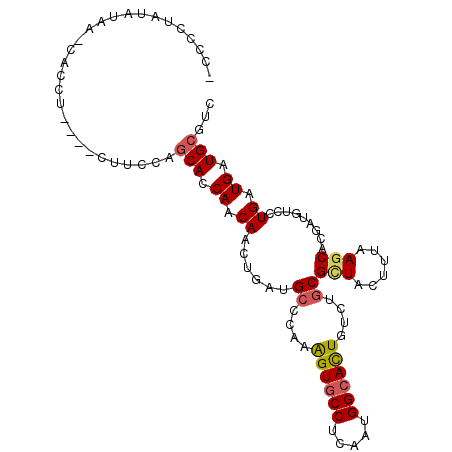
| Location | 21,086,153 – 21,086,252 |
| Length | 99 |
| Max. P | 0.530624 |

| Location | 21,086,153 – 21,086,252 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 77.58 |
| Mean single sequence MFE | -22.48 |
| Consensus MFE | -14.93 |
| Energy contribution | -14.98 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
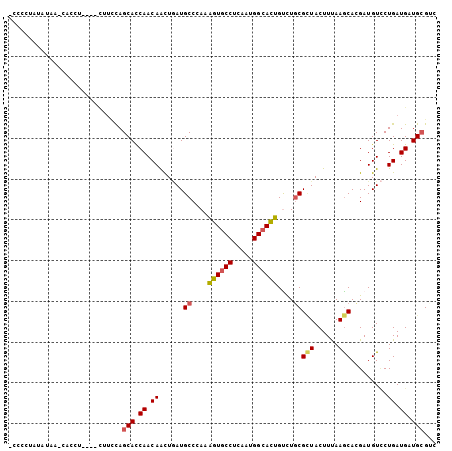
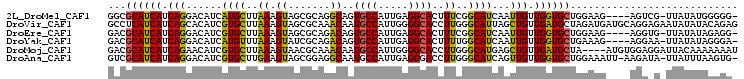
>2L_DroMel_CAF1 21086153 99 + 22407834 -CCCCCAUAUAA-CGACU----CUUCCAGCACCAACAAUUGAUGCCGAAAGUGCCUCAAUGGCACUGCCUGCGCUACUUUAAGCAUGAUGUCCUGAUGAUGCGCC -...........-.....----......(((.((.((...(((((.(..((((((.....))))))..).))(((......))).....))).)).)).)))... ( -21.60) >DroVir_CAF1 9690 105 + 1 CUCUGUAUAUAUUCUCCUGCAUCAUCUAGCAUCAACAGCUAAUGCCCAAGGUGCCCCAAUGGCAUUGUUUGCGCUACUUUAAGCACGAUGUGCUGAUGAUGAGGC ...............(((.(((((((..(((((..(((.(((((((...((....))...))))))).))).(((......)))..)))))...)))))))))). ( -33.00) >DroEre_CAF1 9101 99 + 1 -CCUCUAUAUAA-CACCU----CUUCCAGCACCAACAAUUGAUGCCGAAAGUGCCUCAAUGGCACUGUCUGCGCUACUUUAAGCACGAUGUCCUGAUGAUGCGUC -...........-.....----......(((.((.((((((..((.((.((((((.....)))))).)).))(((......))).))))....)).)).)))... ( -23.10) >DroYak_CAF1 8854 99 + 1 -UCCCUAUAUAA-UUCCU----CUUUCAGCACCAACAAUUGAUGCCAAAAGUGCCUCAAUGGCACUGUCUGCGAUACUUUAAACAUGAUGUCCUGAUGAUGCGUC -...........-.....----......(((.((.((......((....((((((.....))))))....))(((((.........).)))).)).)).)))... ( -17.80) >DroMoj_CAF1 10942 101 + 1 AUUUUUUUGUAAUCCUCCACAU----UAGCAUCAACAGCUCAUGCCCAAGGUGCCCCAAUGGCAUUGUUUGCGUUACUUUAAGCACGAUGUUCUGAUGAUGCGUC ......................----..((((((.(((..(((((....((((((.....))))))....))(((......)))...)))..))).))))))... ( -24.30) >DroAna_CAF1 10297 102 + 1 -CACUUAAAUAA-UAUCUU-AAUUUCCAGCACCAACAACUGAUGCCCAAGGUCCCUCAAUGGCAUUGCCUCCGCUACUUCAAGCACGAUGUCCUGAUGAUGCGAC -...........-......-........(((.((.((...((((((..((....))....))))))......(((......))).........)).)).)))... ( -15.10) >consensus _CCCCUAUAUAA_CACCU____CUUCCAGCACCAACAACUGAUGCCCAAAGUGCCUCAAUGGCACUGUCUGCGCUACUUUAAGCACGAUGUCCUGAUGAUGCGUC ............................(((.((.((......((....((((((.....))))))....))(((......))).........)).)).)))... (-14.93 = -14.98 + 0.06)

| Location | 21,086,153 – 21,086,252 |
|---|---|
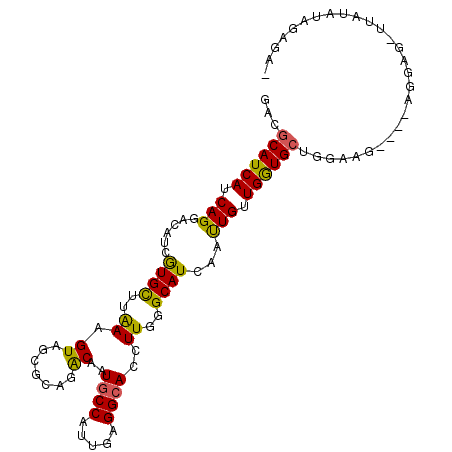
| Length | 99 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 77.58 |
| Mean single sequence MFE | -29.67 |
| Consensus MFE | -17.28 |
| Energy contribution | -16.45 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.58 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21086153 99 - 22407834 GGCGCAUCAUCAGGACAUCAUGCUUAAAGUAGCGCAGGCAGUGCCAUUGAGGCACUUUCGGCAUCAAUUGUUGGUGCUGGAAG----AGUCG-UUAUAUGGGGG- ..(.(((.((.(.(((....(((((..(((.((((.....)))).))).)))))((((((((((((.....))))))))))))----.))).-).))))).)..- ( -31.70) >DroVir_CAF1 9690 105 - 1 GCCUCAUCAUCAGCACAUCGUGCUUAAAGUAGCGCAAACAAUGCCAUUGGGGCACCUUGGGCAUUAGCUGUUGAUGCUAGAUGAUGCAGGAGAAUAUAUACAGAG .((((((((((((((((.((((((......)))))....((((((...((....))...))))))....).)).)))).))))))).)))............... ( -37.00) >DroEre_CAF1 9101 99 - 1 GACGCAUCAUCAGGACAUCGUGCUUAAAGUAGCGCAGACAGUGCCAUUGAGGCACUUUCGGCAUCAAUUGUUGGUGCUGGAAG----AGGUG-UUAUAUAGAGG- ......((.....(((((((((((......))))).....(((((.....)))))(((((((((((.....))))))))))).----.))))-)).....))..- ( -34.20) >DroYak_CAF1 8854 99 - 1 GACGCAUCAUCAGGACAUCAUGUUUAAAGUAUCGCAGACAGUGCCAUUGAGGCACUUUUGGCAUCAAUUGUUGGUGCUGAAAG----AGGAA-UUAUAUAGGGA- ...((.(((...((.(((..(((((..........))))))))))..))).)).((((..((((((.....))))))..))))----.....-...........- ( -23.30) >DroMoj_CAF1 10942 101 - 1 GACGCAUCAUCAGAACAUCGUGCUUAAAGUAACGCAAACAAUGCCAUUGGGGCACCUUGGGCAUGAGCUGUUGAUGCUA----AUGUGGAGGAUUACAAAAAAAU ...((((((.(((....((((((((((.((..(.(((.........))).)..)).)))))))))).))).))))))..----.(((((....)))))....... ( -29.40) >DroAna_CAF1 10297 102 - 1 GUCGCAUCAUCAGGACAUCGUGCUUGAAGUAGCGGAGGCAAUGCCAUUGAGGGACCUUGGGCAUCAGUUGUUGGUGCUGGAAAUU-AAGAUA-UUAUUUAAGUG- ...((((((..(.(((...(((((..(.((..(((.(((...))).)))....)).)..)))))..))).)))))))........-......-...........- ( -22.40) >consensus GACGCAUCAUCAGGACAUCGUGCUUAAAGUAGCGCAGACAAUGCCAUUGAGGCACCUUGGGCAUCAAUUGUUGGUGCUGGAAG____AGGAG_UUAUAUAGAGA_ ...((((((.(((......((((..((.((.......))..((((.....))))..))..))))...))).))))))............................ (-17.28 = -16.45 + -0.83)
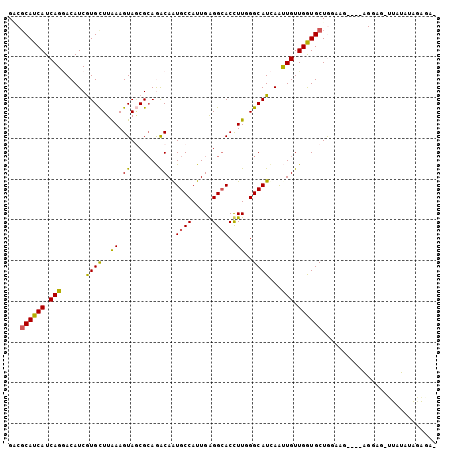
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:41:00 2006