| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,049,652 – 21,049,770 |
| Length | 118 |
| Max. P | 0.970879 |

| Location | 21,049,652 – 21,049,770 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 81.21 |
| Mean single sequence MFE | -25.63 |
| Consensus MFE | -9.91 |
| Energy contribution | -12.65 |
| Covariance contribution | 2.74 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.39 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.970879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
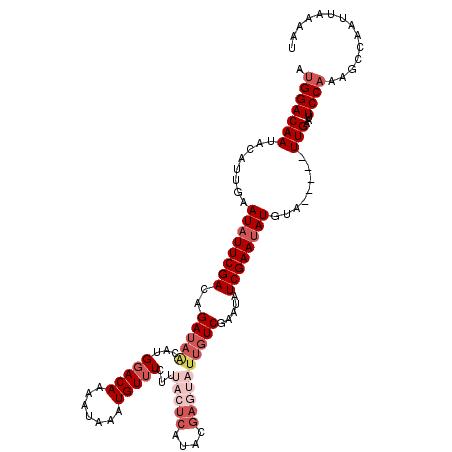
>2L_DroMel_CAF1 21049652 118 + 22407834 AUGGACAAUACAUUGAAUAUUCGACAGAUAACAUGGACAAAAUAAAUGUUUCUUUACUCAUACGAGUAUUGUCGAGUAUCGAAUAUGUAUUGUAUUGAAUUCCAAAGCCAAUUAAAAU .((((((((((((...((((((((((((.(((((...........)))))))..(((((....))))).)))))))))).....))))))))).(((.....)))..)))........ ( -29.80) >DroSec_CAF1 3721 113 + 1 AUGGACAAUACAUUGAAUAUUCGACAGAUAGCACGGACAAAAUAAAUGUUUCUUUACUCAUACGAGUAUUGUCGAAUAUCGAAUAUGUA-----UUGAAUUCCAAAGCCAAUUCAAAU .((((((((((((...(((((((((((.......(((((.......)))))...(((((....)))))))))))))))).....)))))-----)))...)))).............. ( -28.60) >DroSim_CAF1 3672 113 + 1 AUGGACAAUACAUUGAAUAUUCGACAGAUAGCACGGACAAAAUAAAUGUUUCUUUACUCAUACGAGUAUUGUCGAAUAUCGAAUAUGUA-----UUGAAUUCCAAAGCCAAUUCAAAU .((((((((((((...(((((((((((.......(((((.......)))))...(((((....)))))))))))))))).....)))))-----)))...)))).............. ( -28.60) >DroEre_CAF1 3803 107 + 1 AUGGACAAUGCAUUGGAUCUUCGAGAGAUAACGUGGACAAAACAAAUGUUUCUUAACUCAC------AUUGUCGAAUAUCGAAUAUGUA-----UUGAACUCCCAGGGCAAAUAAAAU ........(((.((((...(((((((((((...((.......))..)))))))).....((------((..(((.....)))..)))).-----..)))...)))).)))........ ( -20.00) >DroYak_CAF1 4360 95 + 1 AUGGACAAU----UCGAUAUUCGAGAGAUAACCUGGACAAAUUAAAUGUAUUCCUAC--------------UCGAAUAUCGAAUAUUUA-----UUGUAUUCCAAAGCCAAUUAAAAU .(((.(.((----((((((((((((((.....))(((..............)))..)--------------))))))))))))).....-----(((.....))).))))........ ( -21.14) >consensus AUGGACAAUACAUUGAAUAUUCGACAGAUAACAUGGACAAAAUAAAUGUUUCUUUACUCAUACGAGUAUUGUCGAAUAUCGAAUAUGUA_____UUGAAUUCCAAAGCCAAUUAAAAU .(((((((........((((((((..(((((...(((((.......)))))...(((((....)))))))))).....))))))))........)))...)))).............. ( -9.91 = -12.65 + 2.74)

| Location | 21,049,652 – 21,049,770 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 81.21 |
| Mean single sequence MFE | -24.62 |
| Consensus MFE | -10.05 |
| Energy contribution | -12.81 |
| Covariance contribution | 2.76 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.628457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
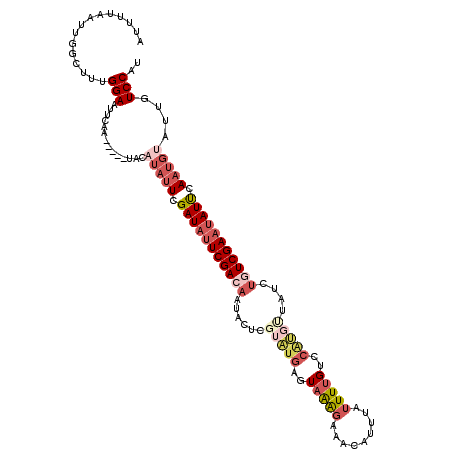
>2L_DroMel_CAF1 21049652 118 - 22407834 AUUUUAAUUGGCUUUGGAAUUCAAUACAAUACAUAUUCGAUACUCGACAAUACUCGUAUGAGUAAAGAAACAUUUAUUUUGUCCAUGUUAUCUGUCGAAUAUUCAAUGUAUUGUCCAU ..............((((...(((((((....((((((((((...((((.(((((....))))).....(((.......)))...))))...))))))))))....))))))))))). ( -26.10) >DroSec_CAF1 3721 113 - 1 AUUUGAAUUGGCUUUGGAAUUCAA-----UACAUAUUCGAUAUUCGACAAUACUCGUAUGAGUAAAGAAACAUUUAUUUUGUCCGUGCUAUCUGUCGAAUAUUCAAUGUAUUGUCCAU ..............((((...(((-----(((((....(((((((((((((((.((...(((((((......)))))))....)).).))).)))))))))))..)))))))))))). ( -28.00) >DroSim_CAF1 3672 113 - 1 AUUUGAAUUGGCUUUGGAAUUCAA-----UACAUAUUCGAUAUUCGACAAUACUCGUAUGAGUAAAGAAACAUUUAUUUUGUCCGUGCUAUCUGUCGAAUAUUCAAUGUAUUGUCCAU ..............((((...(((-----(((((....(((((((((((((((.((...(((((((......)))))))....)).).))).)))))))))))..)))))))))))). ( -28.00) >DroEre_CAF1 3803 107 - 1 AUUUUAUUUGCCCUGGGAGUUCAA-----UACAUAUUCGAUAUUCGACAAU------GUGAGUUAAGAAACAUUUGUUUUGUCCACGUUAUCUCUCGAAGAUCCAAUGCAUUGUCCAU ........(((..((((..(((..-----.......(((.....))).(((------(((.(....(((((....)))))..))))))).......)))..))))..)))........ ( -18.50) >DroYak_CAF1 4360 95 - 1 AUUUUAAUUGGCUUUGGAAUACAA-----UAAAUAUUCGAUAUUCGA--------------GUAGGAAUACAUUUAAUUUGUCCAGGUUAUCUCUCGAAUAUCGA----AUUGUCCAU ..............((((......-----.....(((((((((((((--------------(.(((((((((.......)))....))).)))))))))))))))----))..)))). ( -22.52) >consensus AUUUUAAUUGGCUUUGGAAUUCAA_____UACAUAUUCGAUAUUCGACAAUACUCGUAUGAGUAAAGAAACAUUUAUUUUGUCCAUGUUAUCUGUCGAAUAUUCAAUGUAUUGUCCAU ...............(((..............(((((.(((((((((((......(((((..(((((.........)))))..)))))....))))))))))).)))))....))).. (-10.05 = -12.81 + 2.76)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:55 2006