
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,022,221 – 21,022,351 |
| Length | 130 |
| Max. P | 0.998433 |

| Location | 21,022,221 – 21,022,337 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.47 |
| Mean single sequence MFE | -30.80 |
| Consensus MFE | -28.21 |
| Energy contribution | -28.10 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21022221 116 + 22407834 UAUACAUGUAUGUAAGUAUAUAUCUGGAGGAGGAUUCGUGUGAAAUUUCGUAUGCGGCCUUUGUGUUUGAGUGUUGCGCACACUUGAGUGCACAAUGGGGAAACCUUUUCGCACGC- ....((.((((((....)))))).))..........(((((((((....((..((((((((.......))).)))))((((......))))))...(((....)))))))))))).- ( -32.50) >DroYak_CAF1 78407 106 + 1 ----------UUUAAGUAUAUAUCUGGAGGAGGAUUCGUGUGAAAUUUUGUAUGCGGCCUUUGUGUUUGAGUGCCGCGCACACUUGAGUGCACAAUGGGAAAACCUUUUCGCACGC- ----------...........((((......)))).(((((((((..((((..((((((((.......))).)))))((((......)))))))).((.....)).))))))))).- ( -32.30) >DroAna_CAF1 99113 95 + 1 ----------------------CCUGGAGGAGAAUUCGUGUGAAAUUUUGUAUGCGGCCUUUGUGUUUGAGUGUUGCGCACACUUGAGUGCACAAUGGGGAAGCCUUUUCGCACACA ----------------------((....)).......((((((((..((((..((((((((.......))).)))))((((......)))))))).(((....)))))))))))... ( -27.60) >consensus __________U_UAAGUAUAUAUCUGGAGGAGGAUUCGUGUGAAAUUUUGUAUGCGGCCUUUGUGUUUGAGUGUUGCGCACACUUGAGUGCACAAUGGGGAAACCUUUUCGCACGC_ .........................(((......)))((((((((..((((..((((((((.......))).)))))((((......)))))))).(((....)))))))))))... (-28.21 = -28.10 + -0.11)



| Location | 21,022,221 – 21,022,337 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.47 |
| Mean single sequence MFE | -24.10 |
| Consensus MFE | -20.29 |
| Energy contribution | -19.63 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21022221 116 - 22407834 -GCGUGCGAAAAGGUUUCCCCAUUGUGCACUCAAGUGUGCGCAACACUCAAACACAAAGGCCGCAUACGAAAUUUCACACGAAUCCUCCUCCAGAUAUAUACUUACAUACAUGUAUA -.((((.((((..((((.....((((((((......)))))))).....))))........((....))...)))).))))................(((((..........))))) ( -22.10) >DroYak_CAF1 78407 106 - 1 -GCGUGCGAAAAGGUUUUCCCAUUGUGCACUCAAGUGUGCGCGGCACUCAAACACAAAGGCCGCAUACAAAAUUUCACACGAAUCCUCCUCCAGAUAUAUACUUAAA---------- -.((((.((((.((.....)).((((((((......))))(((((.((.........)))))))..))))..)))).))))..........................---------- ( -26.70) >DroAna_CAF1 99113 95 - 1 UGUGUGCGAAAAGGCUUCCCCAUUGUGCACUCAAGUGUGCGCAACACUCAAACACAAAGGCCGCAUACAAAAUUUCACACGAAUUCUCCUCCAGG---------------------- ((((((((....((.....)).((((((((......)))))))).................))))))))..........................---------------------- ( -23.50) >consensus _GCGUGCGAAAAGGUUUCCCCAUUGUGCACUCAAGUGUGCGCAACACUCAAACACAAAGGCCGCAUACAAAAUUUCACACGAAUCCUCCUCCAGAUAUAUACUUA_A__________ ..((((.((((.((.....)).((((((((......))))))))............................)))).)))).................................... (-20.29 = -19.63 + -0.66)

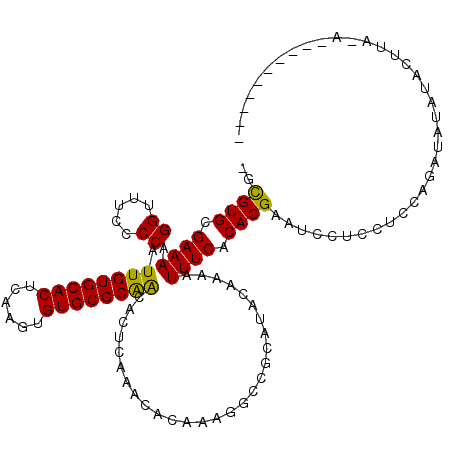
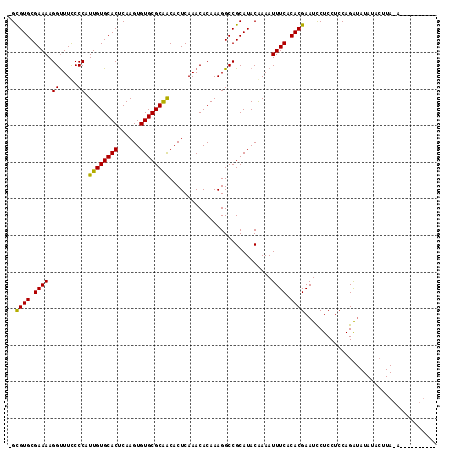
| Location | 21,022,261 – 21,022,351 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 77.78 |
| Mean single sequence MFE | -32.43 |
| Consensus MFE | -23.26 |
| Energy contribution | -22.60 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.72 |
| SVM decision value | 3.10 |
| SVM RNA-class probability | 0.998433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21022261 90 - 22407834 -----------------------UGAGAAUGUGUGUG----GCGUGCGAAAAGGUUUCCCCAUUGUGCACUCAAGUGUGCGCAACACUCAAACACAAAGGCCGCAUACGAAAUUUCA -----------------------.((((.((((((((----((.((.(....((.....)).((((((((......))))))))........).))...))))))))))...)))). ( -29.90) >DroYak_CAF1 78437 90 - 1 -----------------------UGAGAAUGUGUGUG----GCGUGCGAAAAGGUUUUCCCAUUGUGCACUCAAGUGUGCGCGGCACUCAAACACAAAGGCCGCAUACAAAAUUUCA -----------------------.((((.((((((((----((((((.....((.....))..(((((((......))))))))))).....(.....)))))))))))...)))). ( -33.10) >DroAna_CAF1 99131 117 - 1 AUCUGCUGGUUGCUAGUAUGUCCUGUAUAUGUGUGUGUCCUGUGUGCGAAAAGGCUUCCCCAUUGUGCACUCAAGUGUGCGCAACACUCAAACACAAAGGCCGCAUACAAAAUUUCA ...((((((...))))))...........((((((((.(((.((((......((.....)).((((((((......))))))))........)))).))).))))))))........ ( -34.30) >consensus _______________________UGAGAAUGUGUGUG____GCGUGCGAAAAGGUUUCCCCAUUGUGCACUCAAGUGUGCGCAACACUCAAACACAAAGGCCGCAUACAAAAUUUCA .............................((((((((......(((......((.....)).((((((((......))))))))........)))......))))))))........ (-23.26 = -22.60 + -0.66)

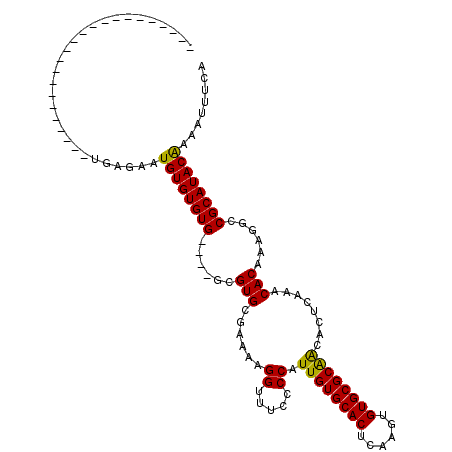
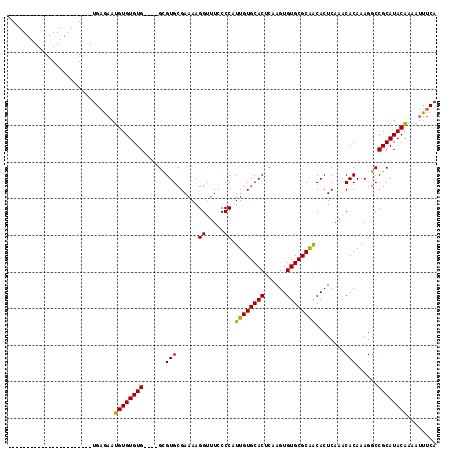
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:51 2006