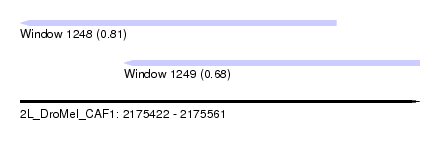
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,175,422 – 2,175,561 |
| Length | 139 |
| Max. P | 0.810471 |

| Location | 2,175,422 – 2,175,532 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 85.71 |
| Mean single sequence MFE | -22.69 |
| Consensus MFE | -13.94 |
| Energy contribution | -14.50 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.810471 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2175422 110 - 22407834 CCCUCGCAC----ACAC--ACACACAGUCAGCCAGCAACUGACUCACUCGCACUCACAUGUGCGAAUCACAACUAACCCGUACAACAACAUCAAAUCAA---CAACUUGGGGCGGCUAA ...((((..----....--......((((((.......))))))...((((((......))))))...........((((...................---.....)))))))).... ( -23.06) >DroEre_CAF1 19235 117 - 1 CCCUCGCACUCACACACACA--UACAGUCGGCCACCAACUGACUCACUCGCACUCACACGUGCGAAUCACAGCCAACCCGAACAACAACAUCAAAUCAACAACAACUUGCGGCGGCAAA ((.(((((............--...((((((.......))))))...((((((......))))))..........................................))))).)).... ( -21.80) >DroYak_CAF1 20029 119 - 1 CCCUCUUACUCACACACACAAACACAGUGGUUCAGCAACUGACUCACUCGCACUCACACGUGCGAAUCACAGCCAACCCGUACAACAACAUCAAAUCAACAACAACUUGCGGCGGCAAA ..........................(((..((((...))))..)))((((((......))))))......(((...(((((.........................))))).)))... ( -23.21) >consensus CCCUCGCACUCACACACACA_ACACAGUCGGCCAGCAACUGACUCACUCGCACUCACACGUGCGAAUCACAGCCAACCCGUACAACAACAUCAAAUCAACAACAACUUGCGGCGGCAAA ...((((..................((((((.......))))))...((((((......))))))..............................................)))).... (-13.94 = -14.50 + 0.56)

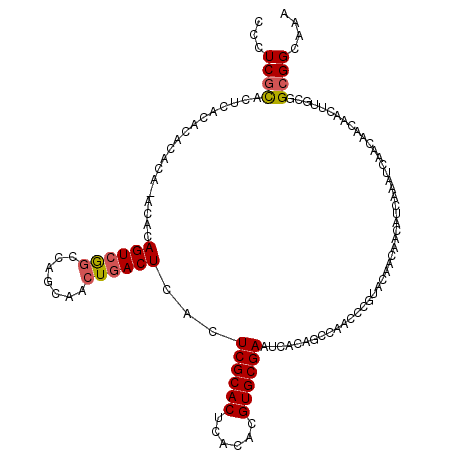
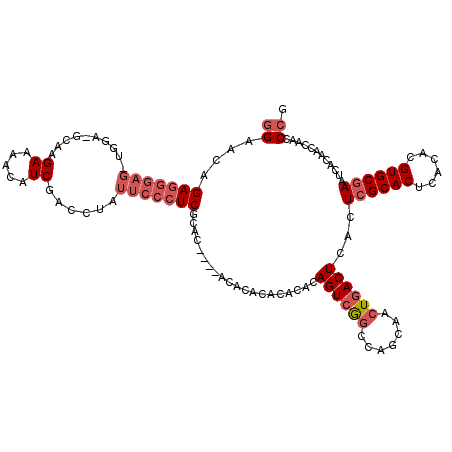
| Location | 2,175,458 – 2,175,561 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.96 |
| Mean single sequence MFE | -27.52 |
| Consensus MFE | -16.83 |
| Energy contribution | -18.47 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.682981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2175458 103 - 22407834 GGAUCAGA-----------GCAAGAAAACAUCGACCUAUUCCCUCGCAC----ACAC--ACACACAGUCAGCCAGCAACUGACUCACUCGCACUCACAUGUGCGAAUCACAACUAACCCG (((..((.-----------(...((.....))..)))..))).......----....--......((((((.......))))))...((((((......))))))............... ( -17.80) >DroSec_CAF1 21499 112 - 1 GGAACAGAGGGAG-UGAA-GGAGGAAAACAUCGACCUAUUCCCUCGCAC----ACAC--ACACACAGUCGGCCAGCAACUGACUCACUCGCACUCACACGUGCGAAUCACAACCAACCCG ((....(((((((-(..(-((..((.....))..)))))))))))....----....--......((((((.......))))))...((((((......)))))).......))...... ( -30.80) >DroSim_CAF1 15869 114 - 1 GGAACAGAGGGAG-UCGA-GCAAGAAAACAUCGACCUAUUCCCUCGCAC----ACACACACACACAGUCGGCCAGCAACUGACUCACUCGCACUCACACGUGCAAAUCACAACCAACCCG ((....(((((((-((((-...........)))))....))))))....----............((((((.......)))))).....((((......)))).........))...... ( -27.80) >DroEre_CAF1 19274 116 - 1 -GAACAGAGGGAGAUGGA-GCAAGAAAACAUCGACCUAUUCCCUCGCACUCACACACACA--UACAGUCGGCCACCAACUGACUCACUCGCACUCACACGUGCGAAUCACAGCCAACCCG -.....(((((((.(((.-(...((.....))..)))))))))))((.............--...((((((.......))))))...((((((......))))))......))....... ( -28.10) >DroYak_CAF1 20068 120 - 1 GGGACUGAGGGAGAUGGAAACAAGAAAACAUCGACCUAUUCCCUCUUACUCACACACACAAACACAGUGGUUCAGCAACUGACUCACUCGCACUCACACGUGCGAAUCACAGCCAACCCG (((...(((((((.((....)).((.....))......))))))).....................(((..((((...))))..)))((((((......))))))...........))). ( -33.10) >consensus GGAACAGAGGGAG_UGGA_GCAAGAAAACAUCGACCUAUUCCCUCGCAC____ACACACACACACAGUCGGCCAGCAACUGACUCACUCGCACUCACACGUGCGAAUCACAACCAACCCG ((....(((((((..........((.....))......)))))))....................((((((.......))))))...((((((......))))))............)). (-16.83 = -18.47 + 1.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:58 2006