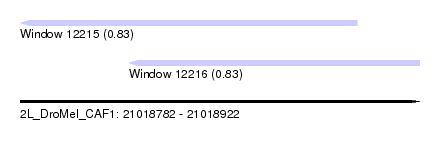
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,018,782 – 21,018,922 |
| Length | 140 |
| Max. P | 0.834724 |

| Location | 21,018,782 – 21,018,900 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 90.96 |
| Mean single sequence MFE | -17.26 |
| Consensus MFE | -14.90 |
| Energy contribution | -14.35 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834444 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21018782 118 - 22407834 AAUAAAACCCAGCAAUUUGUCAAACAAGUUGCUGCCCACGCAGAAAUGAAAUGAAAAAGAAUACAAACCCAACCAGCAAAAUGAAAACAAGAAUUUGUAAAUCUUCCAAAUUAGAAUC .........((((((((((.....))))))))))..((..((....))...))...((((.((((((.........(.....)..........))))))..))))............. ( -15.51) >DroSec_CAF1 64075 118 - 1 AAUAAAACCCAGCAAUUUGUCAAACAAGUUGCUGCCUACGCAGAAAUCAAAUGAAAAAGAAUACAAACACAAACAGCAAAAUGAAAACAAGAAUUUGUAAAUCUUCCAAAUUAGAAUC .........((((((((((.....))))))))))............((.(((....((((.((((((.........(.....)..........))))))..))))....))).))... ( -15.71) >DroEre_CAF1 69917 117 - 1 AAUAAAACCCAGCAAUUUGACAACCAAGUUGCUGCCCACGCAAAAAUCAAGUGAA-AGGAAUGCAAGCCCAAACAGCAAAAUAAAAACAUGAAUUUGUAAAUCUUUCAAAUUAGAAUG .........((((((((((.....)))))))))).................((((-(((..((((((..((..................))..))))))..))))))).......... ( -20.57) >consensus AAUAAAACCCAGCAAUUUGUCAAACAAGUUGCUGCCCACGCAGAAAUCAAAUGAAAAAGAAUACAAACCCAAACAGCAAAAUGAAAACAAGAAUUUGUAAAUCUUCCAAAUUAGAAUC .........((((((((((.....))))))))))............((.(((....((((.((((((..(....................)..))))))..))))....))).))... (-14.90 = -14.35 + -0.55)

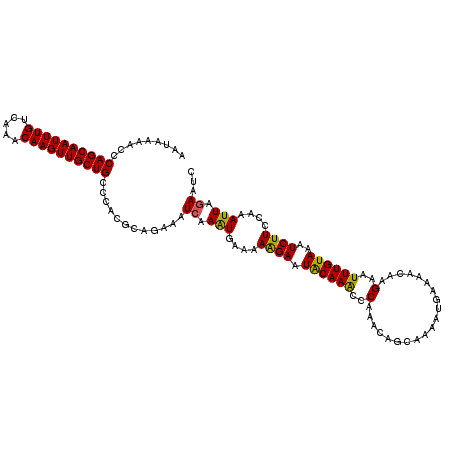

| Location | 21,018,820 – 21,018,922 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 87.58 |
| Mean single sequence MFE | -19.93 |
| Consensus MFE | -15.27 |
| Energy contribution | -15.27 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21018820 102 - 22407834 CAGCCCUCACGCUCUCGGCCCAAAUAAAACCCAGCAAUUUGUCAAACAAGUUGCUGCCCACGCAGAAAUGAAAUGAAAAAGAAUACAAACCCAACCAGCAAA ..........(((...((.............((((((((((.....))))))))))..((..((....))...))...............))....)))... ( -17.30) >DroSec_CAF1 64113 102 - 1 CAGCCCUGAAGCUCUCGGCCCAAAUAAAACCCAGCAAUUUGUCAAACAAGUUGCUGCCUACGCAGAAAUCAAAUGAAAAAGAAUACAAACACAAACAGCAAA ..((.(((..((.....))............((((((((((.....))))))))))......)))...((....)).....................))... ( -16.40) >DroEre_CAF1 69955 101 - 1 AAGCCCUGAAGCUCCGGGCUUAAAUAAAACCCAGCAAUUUGACAACCAAGUUGCUGCCCACGCAAAAAUCAAGUGAA-AGGAAUGCAAGCCCAAACAGCAAA ..........(((..((((((..........((((((((((.....))))))))))..(((...........)))..-........))))))....)))... ( -26.10) >consensus CAGCCCUGAAGCUCUCGGCCCAAAUAAAACCCAGCAAUUUGUCAAACAAGUUGCUGCCCACGCAGAAAUCAAAUGAAAAAGAAUACAAACCCAAACAGCAAA ..(((...........)))............((((((((((.....)))))))))).............................................. (-15.27 = -15.27 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:47 2006