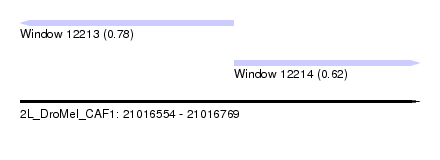
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,016,554 – 21,016,769 |
| Length | 215 |
| Max. P | 0.781926 |

| Location | 21,016,554 – 21,016,669 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.74 |
| Mean single sequence MFE | -31.22 |
| Consensus MFE | -26.93 |
| Energy contribution | -26.93 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781926 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21016554 115 - 22407834 AUUGUAGGUAGCAAGCAUGUAUCUUAAUGAAGGCAAUUUGCUUGUCGAGUUUAGGCAAGUUAGUUUACCGAAAAUUGGCAUAUGGCCAGUAGCAACAA-----AAGCCCCGUACAGGGAA .((((.((((((...(((........)))...))((((.(((((((.......))))))).))))))))....((((((.....)))))).))))...-----....(((.....))).. ( -31.50) >DroPse_CAF1 142848 117 - 1 AUUGUAGGUAGCAAGCAUGUAUCUUAAUGAAGGCAAUUUGCUUGUCGAGUUUAGGCAAGUUAGUUUACCGAAAAUUGGCAUAUGGCCAGUGGCAACAACACAAAAGCAGC-UAC--GCUA .((((.((((((...(((........)))...))((((.(((((((.......))))))).))))))))(...((((((.....))))))..)......)))).(((...-...--))). ( -29.70) >DroEre_CAF1 67668 115 - 1 AUUGUAGGUAGCAAGCAUGUAUCUUAAUGAAGGCAAUUUGCUUGUCGAGUUUAGGCAAGUUAGUUUACCGAAAAUUGGCAUAUGGCCAGUAGCAACAA-----AAGCCCCGUACAGGGCA .((((.((((((...(((........)))...))((((.(((((((.......))))))).))))))))....((((((.....)))))).))))...-----..((((......)))). ( -34.40) >DroYak_CAF1 72688 115 - 1 AUUGUAGGUAGCAAGCAUGUAUCUUAAUGAAGGCAAUUUGCUUGUCGAGUUUAGGCAAGUUAGUUUACCGAAAAUUGGCAUAUGGCCAGUAGCAACAA-----AAGCCCCGUACAGGGCA .((((.((((((...(((........)))...))((((.(((((((.......))))))).))))))))....((((((.....)))))).))))...-----..((((......)))). ( -34.40) >DroAna_CAF1 92505 108 - 1 AUUGUAGGUAGCAAGCAUGUAUCUUAAUGAAGGCAAUUUGCUUGUCGAGUUUAGGCAAGUUAGUUUACCGAAAAUUGGCAUAUGGCCAGUAGCAACAA-----A------GCACAGGAC- .((((.((((((...(((........)))...))((((.(((((((.......))))))).))))))))....((((((.....)))))).((.....-----.------))))))...- ( -27.60) >DroPer_CAF1 135127 117 - 1 AUUGUAGGUAGCAAGCAUGUAUCUUAAUGAAGGCAAUUUGCUUGUCGAGUUUAGGCAAGUUAGUUUACCGAAAAUUGGCAUAUGGCCAGUGGCAACAACACAAAAGCAGC-UAC--GCUA .((((.((((((...(((........)))...))((((.(((((((.......))))))).))))))))(...((((((.....))))))..)......)))).(((...-...--))). ( -29.70) >consensus AUUGUAGGUAGCAAGCAUGUAUCUUAAUGAAGGCAAUUUGCUUGUCGAGUUUAGGCAAGUUAGUUUACCGAAAAUUGGCAUAUGGCCAGUAGCAACAA_____AAGCCCCGUACAGGGCA .((((.((((((...(((........)))...))((((.(((((((.......))))))).))))))))....((((((.....)))))).))))......................... (-26.93 = -26.93 + 0.00)


| Location | 21,016,669 – 21,016,769 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 92.37 |
| Mean single sequence MFE | -28.00 |
| Consensus MFE | -20.66 |
| Energy contribution | -20.34 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624407 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21016669 100 + 22407834 UUAGGCACACAUACGUGCAU--AUAUACAUUGCUAGG--CGUGUGUGUACCUCCGUGGACAUGUAGUGGAACCUACCAUCUUCAUUCAAUGUGCAUUUUACUAG ..(((.(((((((((((((.--........)))...)--))))))))).)))..(((.((((..((((((..........))))))..)))).)))........ ( -27.00) >DroSec_CAF1 61988 102 + 1 UUAGGCACACAUACGUGUAGCCAUAUACAUGGCUAGG--CGUGUGUGUACCUCCGUGGACAUGUAGUGGAACCUACCAUCUUCAUUCAAUGUGCAUUUUACUAG ..(((.((((((((((.(((((((....))))))).)--))))))))).)))..(((.((((..((((((..........))))))..)))).)))........ ( -38.90) >DroSim_CAF1 63815 100 + 1 UUAGGCACACAUACGUGCAU--ACAUACAUGGCUAGG--CGUGUGUGUACCUCCGUGGACAUGUAGUGGAACCUACCAUCUUCAUUCAAUGUGCAUUUUACUAG ....(((((..((((((..(--(((((((((......--))))))))))((.....)).))))))((((......))))..........))))).......... ( -27.60) >DroEre_CAF1 67783 102 + 1 UUAGGCACACAUACGUGCAU--AUAUACAUGGCUAAGGAUGUGUGUGUAUUUCCAUGGACAUGCAGUGGAACCUACCAUCUUCAUUCAAUGUGCAUUUUACUAG ....(((((....((((.((--((((((((..(....)..))))))))))...))))((.(((.(((((......))).)).)))))..))))).......... ( -23.50) >DroYak_CAF1 72803 102 + 1 UUAGGCACACAUACAUGCAU--AUAUACAAGGCUAAGGAUGUGUGUGUAUUUCCAUGGACAUGUAGUGGAACCUACCAUCUUCAUUCAAUGUGCAUUUUACUAG ....(((((....((((.((--(((((((...(....)...)))))))))...))))((.(((.(((((......))).)).)))))..))))).......... ( -23.00) >consensus UUAGGCACACAUACGUGCAU__AUAUACAUGGCUAGG__CGUGUGUGUACCUCCGUGGACAUGUAGUGGAACCUACCAUCUUCAUUCAAUGUGCAUUUUACUAG ....(((((....((((.....(((((((((........))))))))).....))))((.(((.(((((......))).)).)))))..))))).......... (-20.66 = -20.34 + -0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:45 2006