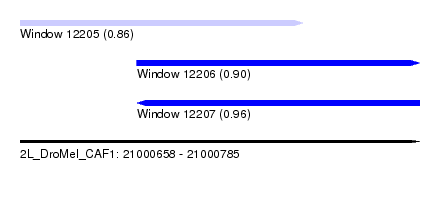
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,000,658 – 21,000,785 |
| Length | 127 |
| Max. P | 0.960424 |

| Location | 21,000,658 – 21,000,748 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 80.24 |
| Mean single sequence MFE | -12.23 |
| Consensus MFE | -11.46 |
| Energy contribution | -11.34 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
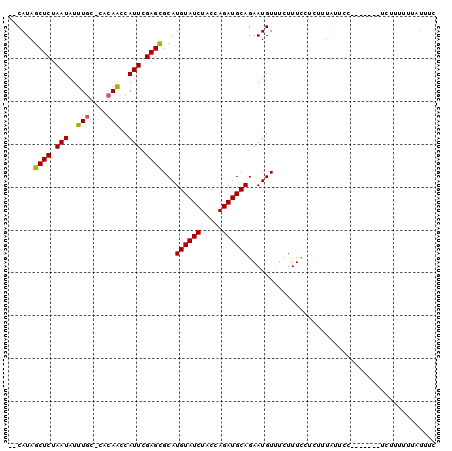
>2L_DroMel_CAF1 21000658 90 + 22407834 --CAUAGCUCUAAUAUUUGC-CACAACCAUUCGAGCGCAUGUAUCUACCAGAUGCAGAAUGUUUCUUUCCUCUUUAUUCC-------UCUUUUUUAUUUC --....((((.(((..(((.-..)))..))).))))...((((((.....))))))(((((.............))))).-------............. ( -12.02) >DroSec_CAF1 49313 90 + 1 --CAUAGCUCUAAUAUUUGC-CACAACCAUUCGAGCGCAUGUAUCUACCAGAUGCAGAAUGUUUCUUUCCUCUUUAUUCC-------UCUUUUUUAUUUC --....((((.(((..(((.-..)))..))).))))...((((((.....))))))(((((.............))))).-------............. ( -12.02) >DroSim_CAF1 49762 90 + 1 --CAUAGCUCUAAUAUUUGC-CACAACCAUUCGAGCGCAUGUAUCUACCAGAUGCAGAAUGUUUCUUUCCUCUUUAUUCC-------UCUUUUUUAUUUC --....((((.(((..(((.-..)))..))).))))...((((((.....))))))(((((.............))))).-------............. ( -12.02) >DroEre_CAF1 55059 83 + 1 --CAUAGCUCUAAUAUUUGC-CACAGCCAUUCGAGCGCAUGUAUCUACCAGAUGCAGAAUGUUUCUUUCCCCUUU-----UUUAUUU---------CUUU --....((((.(((....((-....)).))).))))...((((((.....))))))...................-----.......---------.... ( -11.30) >DroAna_CAF1 80932 76 + 1 AACAUAGCUCUAAUAUUUGCACAGAACCAUUCGAGUGCAUGUAUCUACCAGAUGCAGAAUGUACUUUUUU-UU-----------------------UUUC .((((............(((((.((.....))..)))))((((((.....))))))..))))........-..-----------------------.... ( -13.80) >consensus __CAUAGCUCUAAUAUUUGC_CACAACCAUUCGAGCGCAUGUAUCUACCAGAUGCAGAAUGUUUCUUUCCUCUUUAUUCC_______UCUUUUUUAUUUC ......((((.(((..(((....)))..))).))))...((((((.....))))))............................................ (-11.46 = -11.34 + -0.12)

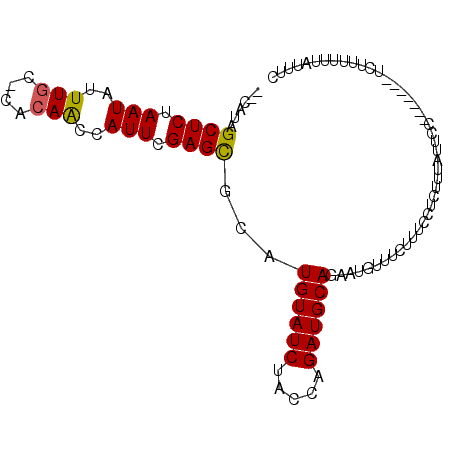
| Location | 21,000,695 – 21,000,785 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 86.63 |
| Mean single sequence MFE | -8.84 |
| Consensus MFE | -8.30 |
| Energy contribution | -8.30 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904821 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21000695 90 + 22407834 GUAUCUACCAGAUGCAGAAUGUUUCUUUCCUCUUUAUUCC-------UCUUUUUUAUUUCUUCCUCUACAGCUUUUCCACAAAUCUGGCUUGCAUUU (((((.....))))).(((((.............))))).-------.......................((....(((......)))...)).... ( -9.02) >DroSec_CAF1 49350 90 + 1 GUAUCUACCAGAUGCAGAAUGUUUCUUUCCUCUUUAUUCC-------UCUUUUUUAUUUCUUCCUCUACAGCUUUUCCACAAAUCUGGCUUGCAUUU (((((.....))))).(((((.............))))).-------.......................((....(((......)))...)).... ( -9.02) >DroSim_CAF1 49799 90 + 1 GUAUCUACCAGAUGCAGAAUGUUUCUUUCCUCUUUAUUCC-------UCUUUUUUAUUUCUUCCUCUACAGCUUUUCCACAAAUCUGGCUUGCAUUU (((((.....))))).(((((.............))))).-------.......................((....(((......)))...)).... ( -9.02) >DroEre_CAF1 55096 83 + 1 GUAUCUACCAGAUGCAGAAUGUUUCUUUCCCCUUU-----UUUAUUU---------CUUUUUCUUCUACAGCUUUUCCACAAAUCUGGCUUGCAUUU (((((.....)))))....................-----.......---------..............((....(((......)))...)).... ( -8.30) >consensus GUAUCUACCAGAUGCAGAAUGUUUCUUUCCUCUUUAUUCC_______UCUUUUUUAUUUCUUCCUCUACAGCUUUUCCACAAAUCUGGCUUGCAUUU (((((.....))))).......................................................((....(((......)))...)).... ( -8.30 = -8.30 + -0.00)

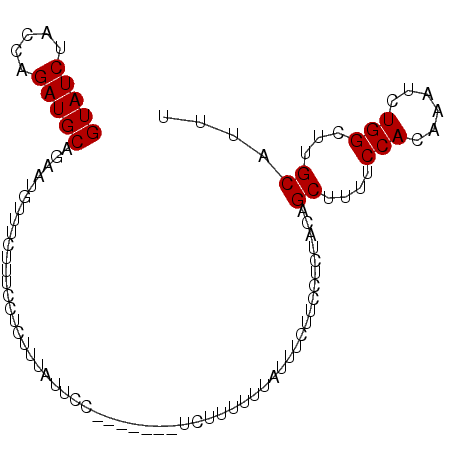
| Location | 21,000,695 – 21,000,785 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 86.63 |
| Mean single sequence MFE | -13.03 |
| Consensus MFE | -12.49 |
| Energy contribution | -12.30 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960424 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21000695 90 - 22407834 AAAUGCAAGCCAGAUUUGUGGAAAAGCUGUAGAGGAAGAAAUAAAAAAGA-------GGAAUAAAGAGGAAAGAAACAUUCUGCAUCUGGUAGAUAC ........((((((..((..(((...((........))............-------..........(........).)))..))))))))...... ( -12.80) >DroSec_CAF1 49350 90 - 1 AAAUGCAAGCCAGAUUUGUGGAAAAGCUGUAGAGGAAGAAAUAAAAAAGA-------GGAAUAAAGAGGAAAGAAACAUUCUGCAUCUGGUAGAUAC ........((((((..((..(((...((........))............-------..........(........).)))..))))))))...... ( -12.80) >DroSim_CAF1 49799 90 - 1 AAAUGCAAGCCAGAUUUGUGGAAAAGCUGUAGAGGAAGAAAUAAAAAAGA-------GGAAUAAAGAGGAAAGAAACAUUCUGCAUCUGGUAGAUAC ........((((((..((..(((...((........))............-------..........(........).)))..))))))))...... ( -12.80) >DroEre_CAF1 55096 83 - 1 AAAUGCAAGCCAGAUUUGUGGAAAAGCUGUAGAAGAAAAAG---------AAAUAAA-----AAAGGGGAAAGAAACAUUCUGCAUCUGGUAGAUAC ........((((((((((..(.....)..))))........---------.......-----...(.((((.......)))).).))))))...... ( -13.70) >consensus AAAUGCAAGCCAGAUUUGUGGAAAAGCUGUAGAGGAAGAAAUAAAAAAGA_______GGAAUAAAGAGGAAAGAAACAUUCUGCAUCUGGUAGAUAC ........((((((..((..(((...((.....))................................(........).)))..))))))))...... (-12.49 = -12.30 + -0.19)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:39 2006