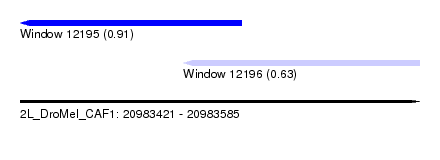
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,983,421 – 20,983,585 |
| Length | 164 |
| Max. P | 0.907626 |

| Location | 20,983,421 – 20,983,512 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 89.23 |
| Mean single sequence MFE | -26.64 |
| Consensus MFE | -24.22 |
| Energy contribution | -23.54 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.907626 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
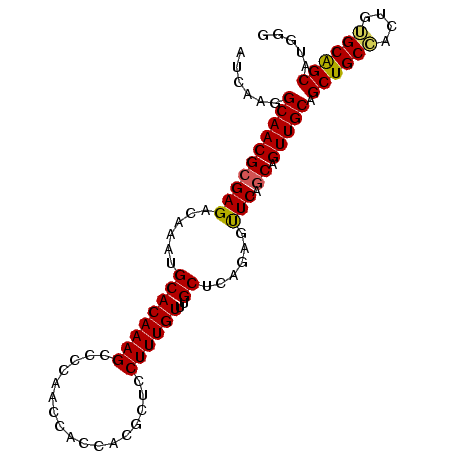
>2L_DroMel_CAF1 20983421 91 - 22407834 GCAGCUGCCACUGUGCAGCAUGGGCAUUGGGAUAUUUUCAAUAUCUAGUAUCGGGUAGCAUACAAAAAACGGUUUGUGCUGAGGGGAUUUA ((.((((((.(.((((.((....))....((((((.....)))))).)))).)))))))..(((((......)))))))............ ( -22.30) >DroSec_CAF1 32268 90 - 1 GCAGCUGCCACUGGGCAGCAUGGGCAUUGGGAUAUUUUCAAUAUCUAGUAUCGGGUAGCACACAAA-AACGGGGUGUGCUGAGGGGAUUUA ...((((((....)))))).(((..((((..(...)..))))..)))..(((...((((((((...-......))))))))....)))... ( -28.80) >DroSim_CAF1 32678 91 - 1 GCAGCUGCCACUGGGCAGCAUGGGCAUUGGGAUAUUUUCAAUAUCUAGUAUCGGGUAGCACACAAAAAACGGGGUGUGCUGAGGGGAUUUA ...((((((....)))))).(((..((((..(...)..))))..)))..(((...((((((((..........))))))))....)))... ( -28.70) >DroEre_CAF1 38526 88 - 1 GCAGCUGCAACUGUGCAGCAU-GGCAAUGGGAUGUUUUCAAUAUCUAGUAUCUGGUGGCACA--GAAGAAGGGGUGUGCUAAGGGGAUUUA ((.((((((....))))))..-.))....((((((.....))))))....(((..(((((((--..........)))))))..)))..... ( -24.50) >DroYak_CAF1 42931 91 - 1 GCAGCUGCAACUGUGCGGCAUUGGCACUGGGAUGUUUUCAAUAUCUAGUAUCUAGUAGCACACAAAAAAAUGGGUGUGCUAAGGGGAUUUA ((.((((((....))))))....))(((((.((((.....)))))))))((((..((((((((..........))))))))...))))... ( -28.90) >consensus GCAGCUGCCACUGUGCAGCAUGGGCAUUGGGAUAUUUUCAAUAUCUAGUAUCGGGUAGCACACAAAAAACGGGGUGUGCUGAGGGGAUUUA ((.((((((....))))))....))....((((((.....))))))...(((...((((((((..........))))))))....)))... (-24.22 = -23.54 + -0.68)


| Location | 20,983,488 – 20,983,585 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 91.62 |
| Mean single sequence MFE | -29.59 |
| Consensus MFE | -24.61 |
| Energy contribution | -24.29 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20983488 97 - 22407834 AUCAAGGCAACGCGAGACAAAUGCACAAAGCCCCAACCACCACGCUCCUUUGUUUGCUCAGAGUUCAGCAGUUGCAGCUGCCACUGUGCAGCAUGGG .(((..(((((((..(((....((((((((.................))))))..)).....)))..)).))))).(((((......))))).))). ( -23.43) >DroSec_CAF1 32334 97 - 1 AUCAAGGCAACGCGAGACAAAUGCACAAAGCCCCAGCCACCACGCUCCUUUGUUUGCUCAGAGCUCAGCAGUUGCAGCUGCCACUGGGCAGCAUGGG .(((..((((((((((......((((((((....(((......))).))))))..))......))).)).))))).((((((....)))))).))). ( -32.00) >DroSim_CAF1 32745 97 - 1 AUCAAGGCAACGCGAGACAAAUGCACAAAGCCCCAGCCACCACGCUCCUUUGUUUGCUCAGAGCUCAGCAGUUGCAGCUGCCACUGGGCAGCAUGGG .(((..((((((((((......((((((((....(((......))).))))))..))......))).)).))))).((((((....)))))).))). ( -32.00) >DroEre_CAF1 38591 93 - 1 AUCAAGGCAACGCGAGACCAGUGCACAAAGCCCCAA---CCACACUGCUUUGUUUGCUCAGAGUUCAGCAGUUGCAGCUGCAACUGUGCAGCAU-GG ......(((((((..((((((((.(((((((.....---.......))))))).))))..).)))..)).))))).((((((....))))))..-.. ( -29.40) >DroYak_CAF1 42998 94 - 1 AUCACGGCAACGCGAGACCAAUGCACAAAGCCCCAA---CCACACUCCUUUGUUUGCUCAGAGUUCAACAGUUGCAGCUGCAACUGUGCGGCAUUGG .((.((....)).))..(((((((.....((.....---........(((((......)))))....((((((((....)))))))))).))))))) ( -31.10) >consensus AUCAAGGCAACGCGAGACAAAUGCACAAAGCCCCAACCACCACGCUCCUUUGUUUGCUCAGAGUUCAGCAGUUGCAGCUGCCACUGUGCAGCAUGGG ......((((((((((......((((((((.................))))))..))......))).)).))))).((((((....))))))..... (-24.61 = -24.29 + -0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:29 2006