
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,979,755 – 20,979,855 |
| Length | 100 |
| Max. P | 0.656586 |

| Location | 20,979,755 – 20,979,855 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 82.44 |
| Mean single sequence MFE | -29.68 |
| Consensus MFE | -17.90 |
| Energy contribution | -20.88 |
| Covariance contribution | 2.99 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.656586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
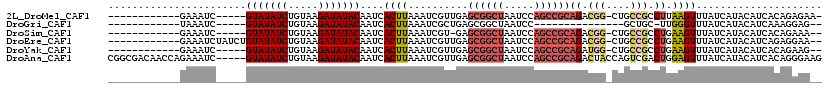
>2L_DroMel_CAF1 20979755 100 + 22407834 -UUCUCUGUGAUGUAUGAUAAACUUAAGGCGGCAG-CCGUCUGCGGCUGGAUUAGCCGCUCAACGAUUUAAGUGAUUGUAUAUCUUACAGAUAUAC-----GAUUUC------------ -...(((((((((((..((..(((((((((....)-))((..(((((((...)))))))...))...)))))).))..))))))..))))).....-----......------------ ( -33.10) >DroGri_CAF1 49003 84 + 1 --CUCCUUUGAUGUAUGAUAAACCCAA-GCAGC---------------GGAUUAGCCGCUCAGCGAUUUAAGUGAUUGUAUAUCUUACAGAUAUAC-----GAUUUA------------ --.........................-(((((---------------((.....)))))..)).........(((((((((((.....)))))))-----))))..------------ ( -19.30) >DroSim_CAF1 28861 98 + 1 --UUUCUGUGAUGUAUGAUAAACUUCAGGCGGCAG-CCGUCUGCGGCUGGAUUAGCCGCUC-ACGAUUUAAGUGAUUGUAUAUCUUACAGAUAUAC-----GAUUUC------------ --....(((((.((.(((......)))(((..(((-(((....)))))).....)))))))-)))........(((((((((((.....)))))))-----))))..------------ ( -32.30) >DroEre_CAF1 34816 104 + 1 --UUCCUCUGAUGUAUGAUAAACUUCAGGCGGCAG-CCGUCUGCGGCUGGAUUAGCCGCUCAACGAUUUAAGUGAUUGUAUAUCUUACAGAUAUACAGAUAGAUUUC------------ --.....((((.((.......)).))))(((((((-(((....)))))......)))))................(((((((((.....))))))))).........------------ ( -29.10) >DroYak_CAF1 38959 99 + 1 --CUUCUGUGAUGUAUGAUAAACUUCAGGCGGCAG-CCAUCUGCGGCUGGAUUAGCCGCUCAACGAUUUAAGUGAUUGUAUAUCUUACAGAUAUAC-----GAUUUC------------ --..(((((((((((..((..((((..(((....)-))....(((((((...)))))))..........)))).))..))))))..))))).....-----......------------ ( -29.60) >DroAna_CAF1 61143 114 + 1 CUUCCCUGUGAUGUAUGAUAAACUCCAGUCGACUGGUAGUCUGCGGCUGGAUUAGCCGCUCAACGAUUUAAGUGAUUGUAUAUCUUACAGAUAUAC-----GAUUUCUGGUUGUCGCCG .......(((((((.(((...(((((((....)))).)))..(((((((...))))))))))))......((.(((((((((((.....)))))))-----)))).))....))))).. ( -34.70) >consensus __UUCCUGUGAUGUAUGAUAAACUUCAGGCGGCAG_CCGUCUGCGGCUGGAUUAGCCGCUCAACGAUUUAAGUGAUUGUAUAUCUUACAGAUAUAC_____GAUUUC____________ .....((((((((((..((..((((.((((((....))))))(((((((...)))))))..........)))).))..))))))..))))............................. (-17.90 = -20.88 + 2.99)


| Location | 20,979,755 – 20,979,855 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 82.44 |
| Mean single sequence MFE | -27.25 |
| Consensus MFE | -14.83 |
| Energy contribution | -16.02 |
| Covariance contribution | 1.18 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20979755 100 - 22407834 ------------GAAAUC-----GUAUAUCUGUAAGAUAUACAAUCACUUAAAUCGUUGAGCGGCUAAUCCAGCCGCAGACGG-CUGCCGCCUUAAGUUUAUCAUACAUCACAGAGAA- ------------....((-----(((((((.....)))))))....((((((...(((..((((((.....)))))).)))((-(....)))))))))...............))...- ( -27.90) >DroGri_CAF1 49003 84 - 1 ------------UAAAUC-----GUAUAUCUGUAAGAUAUACAAUCACUUAAAUCGCUGAGCGGCUAAUCC---------------GCUGC-UUGGGUUUAUCAUACAUCAAAGGAG-- ------------......-----(((((((.....)))))))..((.(((......(..((((((......---------------)))))-)..)((.......))....))))).-- ( -21.10) >DroSim_CAF1 28861 98 - 1 ------------GAAAUC-----GUAUAUCUGUAAGAUAUACAAUCACUUAAAUCGU-GAGCGGCUAAUCCAGCCGCAGACGG-CUGCCGCCUGAAGUUUAUCAUACAUCACAGAAA-- ------------....((-----(((((((.....))))))).............((-((((((((.....))))))....((-(....)))................)))).))..-- ( -27.70) >DroEre_CAF1 34816 104 - 1 ------------GAAAUCUAUCUGUAUAUCUGUAAGAUAUACAAUCACUUAAAUCGUUGAGCGGCUAAUCCAGCCGCAGACGG-CUGCCGCCUGAAGUUUAUCAUACAUCAGAGGAA-- ------------..........((((((((.....))))))))..........(((((..((((((.....)))))).)))))-...((..((((.((.......)).)))).))..-- ( -29.80) >DroYak_CAF1 38959 99 - 1 ------------GAAAUC-----GUAUAUCUGUAAGAUAUACAAUCACUUAAAUCGUUGAGCGGCUAAUCCAGCCGCAGAUGG-CUGCCGCCUGAAGUUUAUCAUACAUCACAGAAG-- ------------....((-----(((((((.....))))))).............(((((((((((.....))))))....((-(....))).........))).))......))..-- ( -24.50) >DroAna_CAF1 61143 114 - 1 CGGCGACAACCAGAAAUC-----GUAUAUCUGUAAGAUAUACAAUCACUUAAAUCGUUGAGCGGCUAAUCCAGCCGCAGACUACCAGUCGACUGGAGUUUAUCAUACAUCACAGGGAAG .((......)).......-----(((((((.....)))))))...........((.(((.((((((.....))))))(((((.((((....)))))))))...........))).)).. ( -32.50) >consensus ____________GAAAUC_____GUAUAUCUGUAAGAUAUACAAUCACUUAAAUCGUUGAGCGGCUAAUCCAGCCGCAGACGG_CUGCCGCCUGAAGUUUAUCAUACAUCACAGAAA__ .......................(((((((.....)))))))....((((..........((((((.....))))))((.(((....))).)).))))..................... (-14.83 = -16.02 + 1.18)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:28 2006