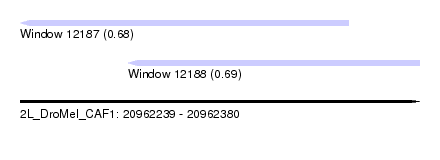
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,962,239 – 20,962,380 |
| Length | 141 |
| Max. P | 0.686329 |

| Location | 20,962,239 – 20,962,355 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.65 |
| Mean single sequence MFE | -24.72 |
| Consensus MFE | -20.52 |
| Energy contribution | -20.27 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
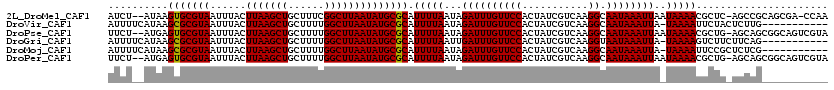
>2L_DroMel_CAF1 20962239 116 - 22407834 AUCU--AUAAGUGCGUAAUUUACUUAAGCUGCUUUCGGCUUAAUAUGCGCAUUUUAAUAGAUUUGUUCCACUAUCGUCAAGGCAAUAAAUUAAUAAAACGCUC-AGCCGCAGCGA-CCAA ((((--(((((((((((...((.((((((((....)))))))))))))))))))..))))))...........((((...(((....................-.)))...))))-.... ( -26.40) >DroVir_CAF1 42567 108 - 1 AUUUUCAUAAGCGCGUAAUUUACUUAAGCUGCUUUUGGCUUAAUAUGCGCAUUUUAAUAGAUUUGUUCCACUAUCGUCAAGGCAAUAAAUUA-UAAAAUUCUACUCUUG----------- ..........(((((((......(((((((......))))))))))))))((((((...((((((((((...........)).)))))))).-))))))..........----------- ( -21.00) >DroPse_CAF1 65597 117 - 1 UUCU--AUGAGUGCGUAAUUUACUUAAGCUGCUUUUGGCUUAAUAUGCGCAUUUUAAUAGAUUUGUUCCACUAUCGUCAAGGCAAUAAAUUAAUAAAACGCUG-AGCAGCGGCAGUCGUA ...(--((((.((((((...)))....(((((((...((.......))((.(((((...((((((((((...........)).))))))))..))))).)).)-)))))).))).))))) ( -29.00) >DroGri_CAF1 35394 108 - 1 AUUUUCAUAAGCGCGUAAUUUACUUAAGCUGCUUUUGGCUUAAUAUGCGCAUUUUAAUUGAUUUGUUCCACUAUCGUCAAGGUAAUAAAUUA-UAAAAGUCUUCUUCAG----------- ..........(((((((......(((((((......)))))))))))))).(((((..(((((((((((...........)).)))))))))-)))))...........----------- ( -21.90) >DroMoj_CAF1 30930 108 - 1 AUUUUCAUAAGCGCGUAAUUUACUUAAGCUGCUUUUGGCUUAAUAUGCGCAUUUUAAUAGAUUUGUUCCACUAUCGUCAAGGCAAUAAAUUA-UAAAAUUCCGCUCUCG----------- ..........(((((((......(((((((......))))))))))))))((((((...((((((((((...........)).)))))))).-))))))..........----------- ( -21.00) >DroPer_CAF1 55623 117 - 1 UUCU--AUGAGUGCGUAAUUUACUUAAGCUGCUUUUGGCUUAAUAUGCGCAUUUUAAUAGAUUUGUUCCACUAUCGUCAAGGCAAUAAAUUAAUAAAACGCUG-AGCAGCGGCAGUCGUA ...(--((((.((((((...)))....(((((((...((.......))((.(((((...((((((((((...........)).))))))))..))))).)).)-)))))).))).))))) ( -29.00) >consensus AUCU__AUAAGCGCGUAAUUUACUUAAGCUGCUUUUGGCUUAAUAUGCGCAUUUUAAUAGAUUUGUUCCACUAUCGUCAAGGCAAUAAAUUA_UAAAACGCUG_AGCAG___________ ..........(((((((......(((((((......)))))))))))))).(((((...((((((((((...........)).))))))))..)))))...................... (-20.52 = -20.27 + -0.25)
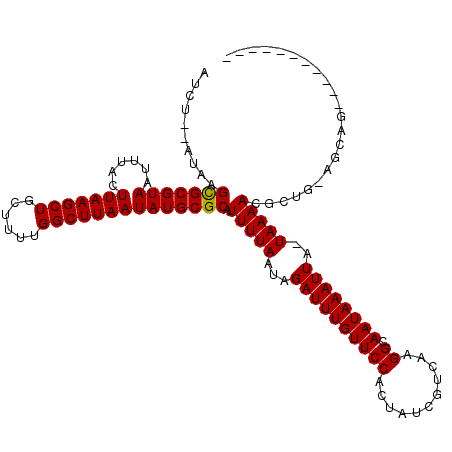
| Location | 20,962,277 – 20,962,380 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.58 |
| Mean single sequence MFE | -20.85 |
| Consensus MFE | -15.27 |
| Energy contribution | -15.02 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686329 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20962277 103 - 22407834 --AG-------------CCCGAUUCGUUUUCGUUAUUGCCAUCU--AUAAGUGCGUAAUUUACUUAAGCUGCUUUCGGCUUAAUAUGCGCAUUUUAAUAGAUUUGUUCCACUAUCGUCAA --..-------------..((((.((....))........((((--(((((((((((...((.((((((((....)))))))))))))))))))..))))))..........)))).... ( -20.80) >DroVir_CAF1 42595 98 - 1 ----------------------GUUGUCUUCGUUUUUUCCAUUUUCAUAAGCGCGUAAUUUACUUAAGCUGCUUUUGGCUUAAUAUGCGCAUUUUAAUAGAUUUGUUCCACUAUCGUCAA ----------------------............................(((((((......(((((((......)))))))))))))).............................. ( -15.80) >DroPse_CAF1 65636 116 - 1 --GGACUCUGACUCUCCUCCGAUUCUCCUUCUUGGUCGCAUUCU--AUGAGUGCGUAAUUUACUUAAGCUGCUUUUGGCUUAAUAUGCGCAUUUUAAUAGAUUUGUUCCACUAUCGUCAA --.(((...((.((......)).)).......(((..(((.(((--(((((((((((...((.(((((((......))))))))))))))))))..)))))..))).))).....))).. ( -26.70) >DroGri_CAF1 35422 98 - 1 ----------------------GUCGCCUUUGUUCUUUUCAUUUUCAUAAGCGCGUAAUUUACUUAAGCUGCUUUUGGCUUAAUAUGCGCAUUUUAAUUGAUUUGUUCCACUAUCGUCAA ----------------------(.((....((..(...(((.((......(((((((......(((((((......)))))))))))))).....)).)))...)...))....)).).. ( -16.00) >DroMoj_CAF1 30958 98 - 1 ----------------------GUUGGCUUCGUUCUUUCCAUUUUCAUAAGCGCGUAAUUUACUUAAGCUGCUUUUGGCUUAAUAUGCGCAUUUUAAUAGAUUUGUUCCACUAUCGUCAA ----------------------.(((((......................(((((((......(((((((......)))))))))))))).........(((..........)))))))) ( -19.10) >DroAna_CAF1 50081 105 - 1 GUGG-------------CUUGAUUCGGUUUCGUUAUUGCCAUCU--AUAAGUGCGUAAUUUACUUAAGCUGCUUUCGGCUUAAUAUGCGCAUUUUAAUAGAUUUGUUCCACUAUCGUCAA ((((-------------..(((...(((.........)))((((--(((((((((((...((.((((((((....)))))))))))))))))))..)))))))))..))))......... ( -26.70) >consensus ______________________GUCGCCUUCGUUAUUGCCAUCU__AUAAGCGCGUAAUUUACUUAAGCUGCUUUUGGCUUAAUAUGCGCAUUUUAAUAGAUUUGUUCCACUAUCGUCAA ..................................................(((((((......(((((((......)))))))))))))).............................. (-15.27 = -15.02 + -0.25)

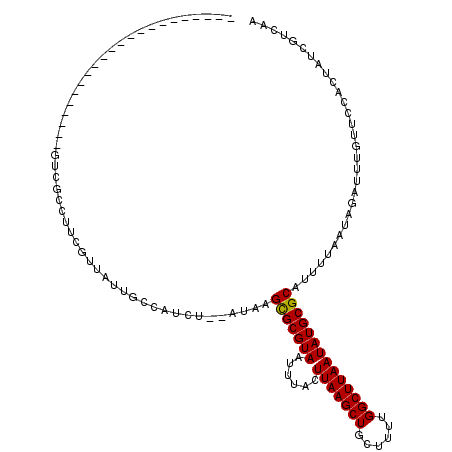
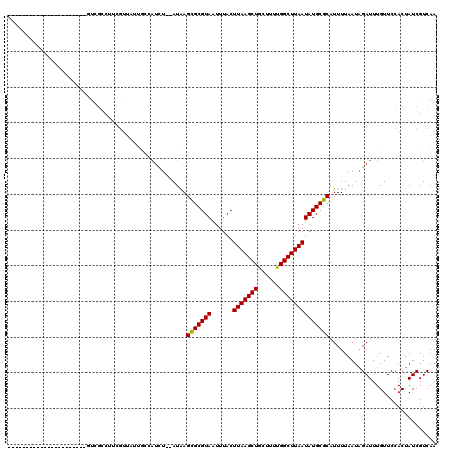
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:22 2006