
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
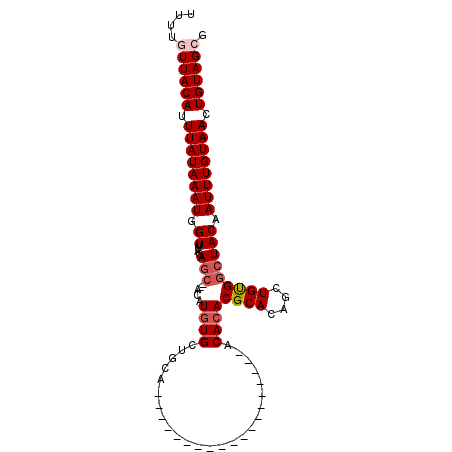
| Location | 20,956,370 – 20,956,464 |
| Length | 94 |
| Max. P | 0.742444 |

| Location | 20,956,370 – 20,956,464 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 81.24 |
| Mean single sequence MFE | -22.19 |
| Consensus MFE | -13.25 |
| Energy contribution | -13.86 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.712506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
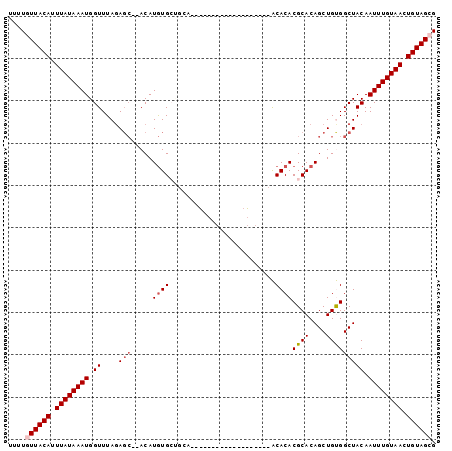
>2L_DroMel_CAF1 20956370 94 + 22407834 UUUUGUUACAUUUAUAAAUGGUUUAGAGCGCACAUGUGCCGCACAG-------CACCAAAACACACACGCACAGCUGUGGCUACAAUUUGUAACUGUAGCG ....((((((.((((((((.((....((((((....)))..(((((-------(...................))))))))))).)))))))).)))))). ( -25.51) >DroVir_CAF1 34996 99 + 1 UUUUGUUACAUUUAUAAAUGGUUUAGAGC--ACAUGUGCUGUACACACACAGACACGAGCACACACACGCACAGCUGUGGCUACAAUUUGUAACUGUAGCG ....((((((.((((((((.((.((.(((--(....)))).)).)).(((((....(.((........)).)..)))))......)))))))).)))))). ( -26.80) >DroPse_CAF1 58163 75 + 1 UUUUGUUACAUUUAUAAAUGGUUUAGAGC--ACAUGUG------------------------GCACACGCACAGCUGCGGCUACAAUUUGUAACUGUAGAG .....(((((.((((((((.((.....))--...((((------------------------((...((((....)))))))))))))))))).))))).. ( -18.30) >DroGri_CAF1 28768 81 + 1 UUUUGUUACAUUUAUAAAUGGUUUACACA--ACAUAUGCUACAA------------------ACACACGCACAGCUGUGGCUACAAUUUGUAACUGUAGCG ....((((((.((((((((.((...((((--.(...(((.....------------------......)))..).))))...)).)))))))).)))))). ( -19.00) >DroAna_CAF1 44480 86 + 1 UUUUGUUACAUUUAUAAAUGGUUUAGAGCACACAUGUGCUGCCU---------------CACACACACGCACAGCUGUGGCUACAAUUUGUAACUGUAGCG ....((((((.((((((((.((....(((.(((((((((((...---------------......)).)))))..))))))))).)))))))).)))))). ( -25.20) >DroPer_CAF1 48057 75 + 1 UUUUGUUACAUUUAUAAAUGGUUUAGAGC--ACAUGUG------------------------GCACACGCACAGCUGCGGCUACAAUUUGUAACUGUAGAG .....(((((.((((((((.((.....))--...((((------------------------((...((((....)))))))))))))))))).))))).. ( -18.30) >consensus UUUUGUUACAUUUAUAAAUGGUUUAGAGC__ACAUGUGCUGCA___________________ACACACGCACAGCUGUGGCUACAAUUUGUAACUGUAGCG ....((((((.((((((((.((....(((.....((((.........................))))((((....))))))))).)))))))).)))))). (-13.25 = -13.86 + 0.61)

| Location | 20,956,370 – 20,956,464 |
|---|---|
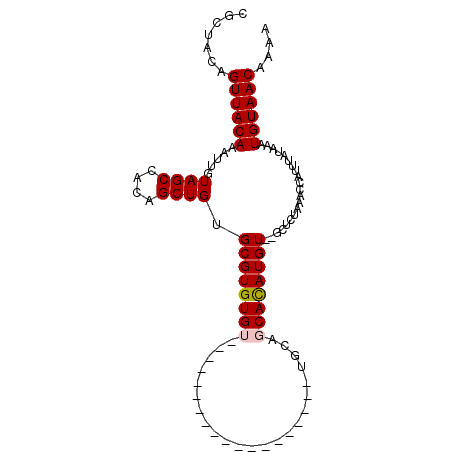
| Length | 94 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 81.24 |
| Mean single sequence MFE | -21.40 |
| Consensus MFE | -14.51 |
| Energy contribution | -14.70 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742444 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
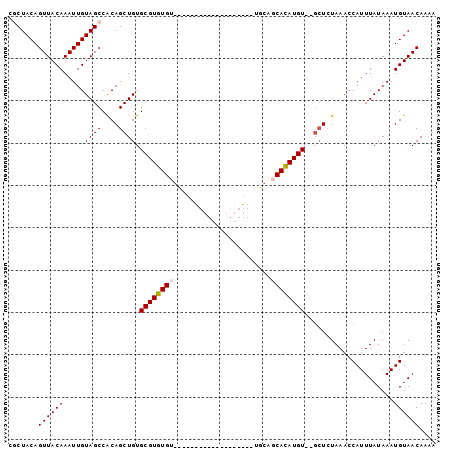
>2L_DroMel_CAF1 20956370 94 - 22407834 CGCUACAGUUACAAAUUGUAGCCACAGCUGUGCGUGUGUGUUUUGGUG-------CUGUGCGGCACAUGUGCGCUCUAAACCAUUUAUAAAUGUAACAAAA .((((((((.....))))))))......(((((((.((((...((((.-------..(.(((.((....))))).)...))))..)))).)))).)))... ( -25.90) >DroVir_CAF1 34996 99 - 1 CGCUACAGUUACAAAUUGUAGCCACAGCUGUGCGUGUGUGUGCUCGUGUCUGUGUGUGUACAGCACAUGU--GCUCUAAACCAUUUAUAAAUGUAACAAAA .((((((((.....)))))))).(((..((((.(((.((..((.((((((((((....))))).))))).--)).....))))).))))..)))....... ( -28.20) >DroPse_CAF1 58163 75 - 1 CUCUACAGUUACAAAUUGUAGCCGCAGCUGUGCGUGUGC------------------------CACAUGU--GCUCUAAACCAUUUAUAAAUGUAACAAAA .......((((((......(((....)))(..((((...------------------------..)))).--.).................)))))).... ( -15.20) >DroGri_CAF1 28768 81 - 1 CGCUACAGUUACAAAUUGUAGCCACAGCUGUGCGUGUGU------------------UUGUAGCAUAUGU--UGUGUAAACCAUUUAUAAAUGUAACAAAA .((((((((.....))))))))(((((((((((......------------------.....))))).))--))))......................... ( -21.10) >DroAna_CAF1 44480 86 - 1 CGCUACAGUUACAAAUUGUAGCCACAGCUGUGCGUGUGUGUG---------------AGGCAGCACAUGUGUGCUCUAAACCAUUUAUAAAUGUAACAAAA .((((((((.....)))))))).......(..((..(((((.---------------.....)))))..))..)........................... ( -22.80) >DroPer_CAF1 48057 75 - 1 CUCUACAGUUACAAAUUGUAGCCGCAGCUGUGCGUGUGC------------------------CACAUGU--GCUCUAAACCAUUUAUAAAUGUAACAAAA .......((((((......(((....)))(..((((...------------------------..)))).--.).................)))))).... ( -15.20) >consensus CGCUACAGUUACAAAUUGUAGCCACAGCUGUGCGUGUGU___________________UGCAGCACAUGU__GCUCUAAACCAUUUAUAAAUGUAACAAAA .......((((((.....((((....)))).((((((((.......................)))))))).....................)))))).... (-14.51 = -14.70 + 0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:20 2006