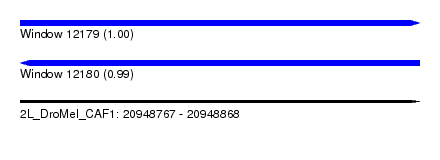
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,948,767 – 20,948,868 |
| Length | 101 |
| Max. P | 0.995779 |

| Location | 20,948,767 – 20,948,868 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 70.64 |
| Mean single sequence MFE | -35.22 |
| Consensus MFE | -20.30 |
| Energy contribution | -20.97 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.58 |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.995779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
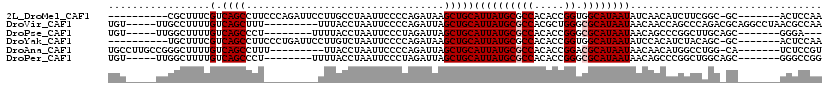
>2L_DroMel_CAF1 20948767 101 + 22407834 UUGGAGU-------GC-GCCGAAGAUGUUGAUAUUAUGCCACCGGUGUGGCGCAUAAUGCAGCUUAUCUGGGGAAUUAGGCAAGGAAUCUGGGAAGGCUGACGAAAGCG---------- .....((-------((-(((...((((....))))(((((...))))))))))))....((((((.((..((...((.....))...))..)).)))))).(....)..---------- ( -25.70) >DroVir_CAF1 21575 105 + 1 UUGGCGUUAGGCCUGCGUCUGGGCUGGUUGUUAUUAUGCGCCCAGCGUGGCGCAUAAUGCAGCUAAUCUGGGGAAUUAGGUAAA---------AAAGCUGACAAAAGGCAA-----ACA (..((.....(((((..((..(..(((((((.((((((((((......)))))))))))))))))..)..))....)))))...---------...))..)..........-----... ( -38.70) >DroPse_CAF1 43641 96 + 1 ---UCCC-------GCUGCAAGCCGGGCUGUUAUUAUGCGCCCGGUGUGGCGCAUAAUGCAGCUAAUCUAGGGAAUUAGGUAAAA--------AGGGCUGACAAAAGCCAA-----ACA ---((((-------((.....))..((((((.((((((((((......))))))))))))))))......))))...........--------..((((......))))..-----... ( -38.10) >DroYak_CAF1 13134 101 + 1 UUGGAGU-------GC-GCUGUAGAUGUGGAUAUUAUGCCACCGGUGUGGCGCAUAAUGCAGCUUAUCUGGGGAAUUAGACAAGGAAUCAGGGAAGGCUGACGAAAGCA---------- .....((-------((-((..((...((((........))))...))..))))))....((((((.(((((....((....))....)))))..)))))).(....)..---------- ( -28.80) >DroAna_CAF1 37066 102 + 1 ACGGAGA-------UG-CCAGGCCAUGUUGUUAUUAUGCGUCCGGUGUGGCGCAUAAUGCAGCUAAUCUGGGGAAUUAGGUAA---------AAAGGCUGACAAAAGCCCGGCAAGGCA .......-------((-((..(((..(((((.((((((((((......)))))))))))))))..((((((....))))))..---------...((((......)))).)))..)))) ( -37.90) >DroPer_CAF1 34602 99 + 1 CCGGCCC-------GCUGCCAGCCGGGCUGUUAUUAUGCGCCCGGUGUGGCGCAUAAUGCAGCUAAUCUAGGGAAUUAGGUAAAA--------AGGGCUGACAAAAGCCAA-----ACA ..(((..-------..((.(((((.((((((.((((((((((......)))))))))))))))).((((((....))))))....--------..))))).))...)))..-----... ( -42.10) >consensus UUGGAGU_______GC_GCAGGCCAGGUUGUUAUUAUGCGCCCGGUGUGGCGCAUAAUGCAGCUAAUCUGGGGAAUUAGGUAAA_________AAGGCUGACAAAAGCCAA_____ACA ..........................((((.(((((((((((......)))))))))))))))..((((((....))))))...............(((......)))........... (-20.30 = -20.97 + 0.67)

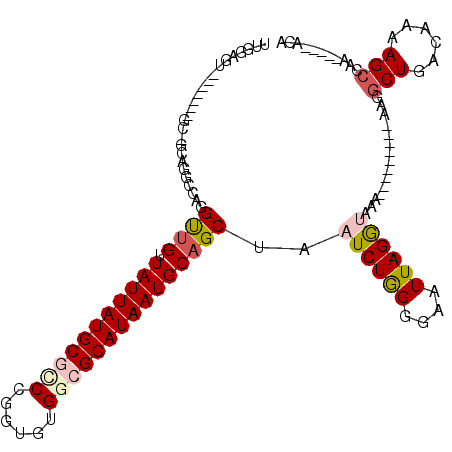
| Location | 20,948,767 – 20,948,868 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 70.64 |
| Mean single sequence MFE | -29.61 |
| Consensus MFE | -11.71 |
| Energy contribution | -12.04 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.40 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.994027 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20948767 101 - 22407834 ----------CGCUUUCGUCAGCCUUCCCAGAUUCCUUGCCUAAUUCCCCAGAUAAGCUGCAUUAUGCGCCACACCGGUGGCAUAAUAUCAACAUCUUCGGC-GC-------ACUCCAA ----------...........(((..............((.((((....(((.....))).)))).))(((((....))))).................)))-..-------....... ( -17.50) >DroVir_CAF1 21575 105 - 1 UGU-----UUGCCUUUUGUCAGCUUU---------UUUACCUAAUUCCCCAGAUUAGCUGCAUUAUGCGCCACGCUGGGCGCAUAAUAACAACCAGCCCAGACGCAGGCCUAACGCCAA .((-----(.((((..((((.(((..---------.....((((((.....))))))....((((((((((......)))))))))).......)))...)))).))))..)))..... ( -29.40) >DroPse_CAF1 43641 96 - 1 UGU-----UUGGCUUUUGUCAGCCCU--------UUUUACCUAAUUCCCUAGAUUAGCUGCAUUAUGCGCCACACCGGGCGCAUAAUAACAGCCCGGCUUGCAGC-------GGGA--- ...-----(((((....)))))(((.--------......(((......)))....(((((((((((((((......))))))))))...(((...))).)))))-------))).--- ( -33.20) >DroYak_CAF1 13134 101 - 1 ----------UGCUUUCGUCAGCCUUCCCUGAUUCCUUGUCUAAUUCCCCAGAUAAGCUGCAUUAUGCGCCACACCGGUGGCAUAAUAUCCACAUCUACAGC-GC-------ACUCCAA ----------(((....(((((......)))))..(((((((........)))))))..)))...(((((.......((((........)))).......))-))-------)...... ( -23.04) >DroAna_CAF1 37066 102 - 1 UGCCUUGCCGGGCUUUUGUCAGCCUUU---------UUACCUAAUUCCCCAGAUUAGCUGCAUUAUGCGCCACACCGGACGCAUAAUAACAACAUGGCCUGG-CA-------UCUCCGU .....(((((((((.((((........---------....((((((.....))))))....((((((((((.....)).)))))))).))))...)))))))-))-------....... ( -32.10) >DroPer_CAF1 34602 99 - 1 UGU-----UUGGCUUUUGUCAGCCCU--------UUUUACCUAAUUCCCUAGAUUAGCUGCAUUAUGCGCCACACCGGGCGCAUAAUAACAGCCCGGCUGGCAGC-------GGGCCGG ...-----.(((((((((((((((..--------......(((......)))....((((.((((((((((......))))))))))..))))..))))))))).-------)))))). ( -42.40) >consensus UGU_____UUGGCUUUUGUCAGCCUU_________UUUACCUAAUUCCCCAGAUUAGCUGCAUUAUGCGCCACACCGGGCGCAUAAUAACAACAUGGCCAGC_GC_______ACGCCAA .................(.((((.................................)))))((((((((((.....)).))))))))................................ (-11.71 = -12.04 + 0.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:40:15 2006